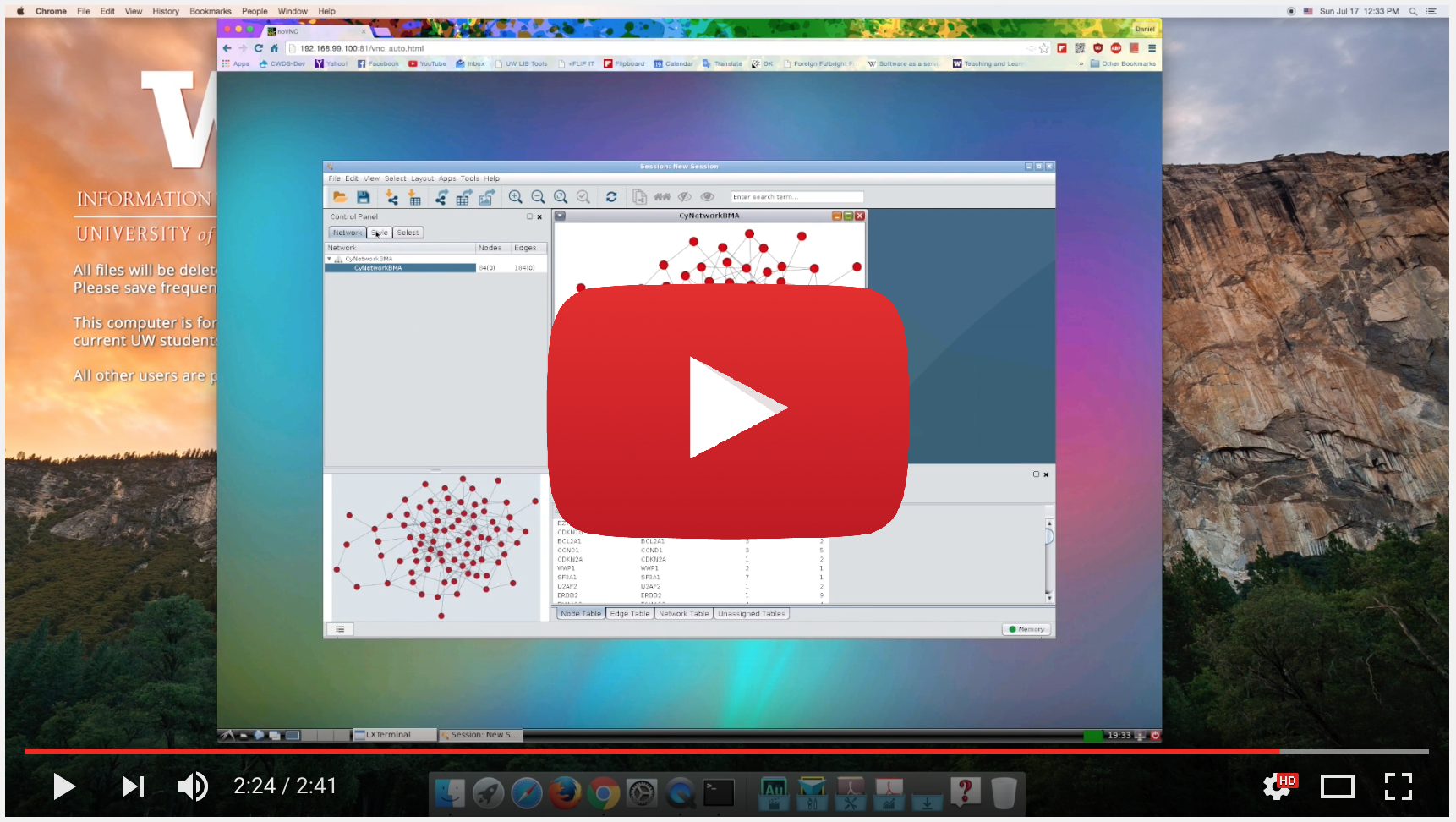
The purpose of this project is to address the issue of delivering graphical interface from Docker container. Our approach is by using the noVNC solution, developed by Kanaka et.all.
Basically, noVNC is to use browser as VNC client to the running container, giving more freedom and flexibility on having graphical applications running as a docker package.
This package will get you: Cytoscape (a open source java application with GUI), R and Bioconductor, and CyNetworkBMA (a Cytoscape plug in to infer gene networks).
GUIdock-VNC: Using a graphical desktop sharing system to provide a browser-based interface for containerized software. Varun Mittal, Ling-Hong Hung, Jayant Keswani, Daniel Kristiyanto, Sung Bong Lee and Ka Yee Yeung. Gigascience 2017, 6(4): 1-6. (https://academic.oup.com/gigascience/article/3052724/GUIdock-VNC:)
A more detailed on how to get running is available here
CyNetworkBMA is a cytoscape plugin to infer gene networks using bayesian techniques. A more information about CynetworkBMA is available here
This container was tested on various operating systems (MacOS, Windows, Linux) and various environtments (laptop/desktop, Microsoft Azure Cloud, Google Cloud, and AWS).
In any docker-enabled environtment, the container can be run using following commands:
# Make sure to use the latest image
docker pull biodepot/novnc-cynetworkbma
#
docker run -v /Users/path/to/your/files:/data -d -p 80:6080 biodepot/novnc-cynetworkbma
Once run, use your favorite browser to interact with the container. To get the IP address of your docker, use following command:
# List of running docker machines
docker-machine ps
# Get the docker machine IP
docker-machine ip default
You are invited to post a questions or sugesstion by pushing push request or sending 'issues' on this repo.