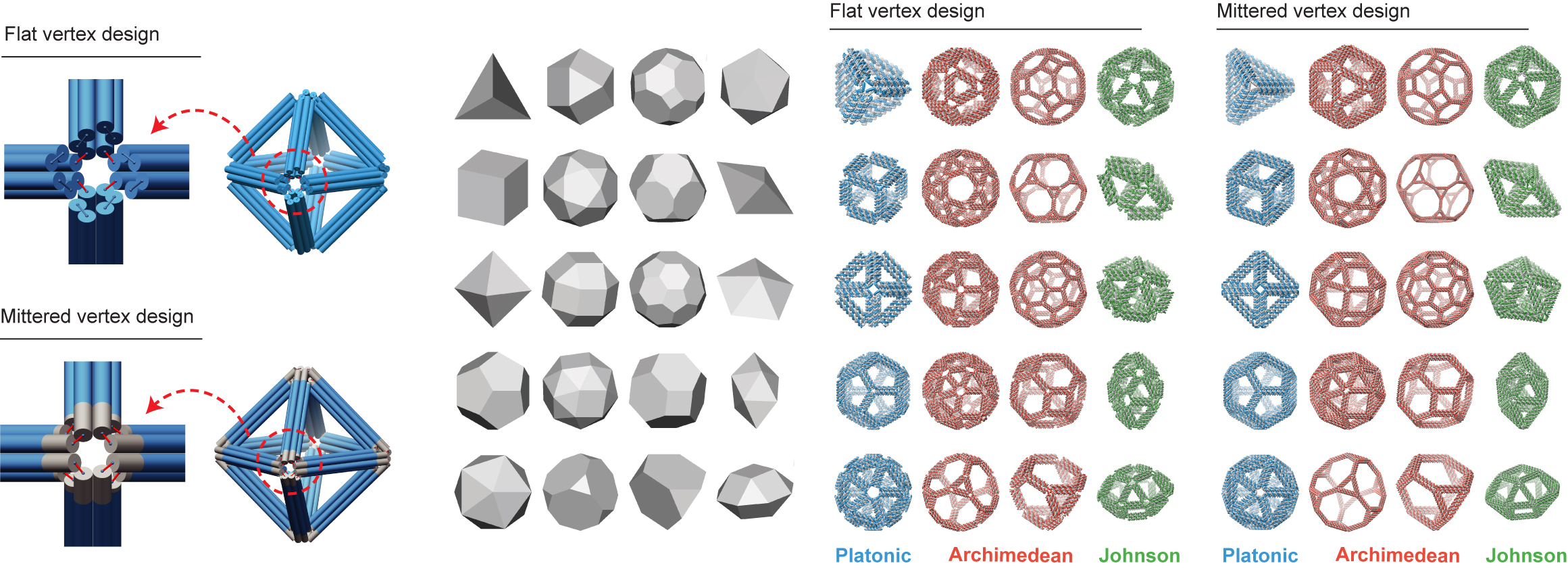
TALOS (Three-dimensional, Algorithmically-generated Library of DNA Origami Shapes) is a new, free and open-source software package written by FORTRAN 90/95 that enables the automated convertion of 3D computer-generated design files (PLY) into DNA sequences. These DNA sequences can be subsequently synthesized and mixed to fold DNA 6HB based wireframe 3D nanoparticles with high fidelity.
Documentation:
The full software documention can be downloaded here.
The *.csv file generated by TALOS contains final staple sequences.
The *.bild and *.json generated by TALOS can be opened by UCSF Chimera and caDNAno.
The atomic model (PDB) can be generated by *.cndo file using the atomic model generator written by Dr. Keyao Pan.
In addition to the source code and pre-compiled binaries available here, we also offer an online web application with all the functionality found in the downloaded version. By submitting the same inputs (PLY), the web service will return the same file output as the downloaded version.
git clone https://github.com/hmjeon/TALOS.git
Requirements to compile from source:
Intel Fortran compiler: Intel Parallel Studio XE 2016, 2017 or 2018
- Compiling the TALOS sources require Intel Fortran. Free Intel (R) Software Development Tools are available for qualified students, educators, academic researchers and open source contributors, see the details.
- The Intel Fortran compiler supports all of the features of the Fortran 90, Fortran 95, Fortran 2003 standards and most of Fortran 2008. It also supports some draft Fortran 2018 features.
- We provide MakeFile which is a simple way to organize code compilation of TALOS.
- Fully automatic procedure of the sequence design for scaffolded DNA 6HB-based wireframe nanoparticles
- Importing PLY file format as an input
- Two vertex designs: flat and mitered vertices
- JSON output for editing staple paths and sequences from caDNAno
- 3D visual outputs by UCSF Chimera
- A library of 240 nanoparticles - 40 geometries, 2 vertex designs, and 3 different edge lengths
- User-friendly TUI (Text-based User Interface)
- Online web resources and release packages for Microsoft Windows and Mac OS
- Free and open source (GNU General Public License, version 3.0)
Dr. Hyungmin Jun (hyungminjun@outlook.com), LCBB (Laboratory of Computational Biology and Biophysics), MIT
PERDIX is an open-source software distributed under the GPL license, version 3