- A Kishore Kumar
- Vidit Jain
This project involves optimizing a dynamic programming algorithm that cannot be parallelized trivially.
In this project, we worked on reordering the way we process the recurrence relation by iterating over the diagonals instead of a row-wise or column-wise method, and stored only the diagonals needed for computing the future values to save space. Doing so allowed us to make the algorithm parallelizable by removing inter-dependencies.
We also modified the way we stored these diagonals by storing them in a linear fashion instead of accessing the elements as a diagonal of a grid, making the array access much more cache-efficient.
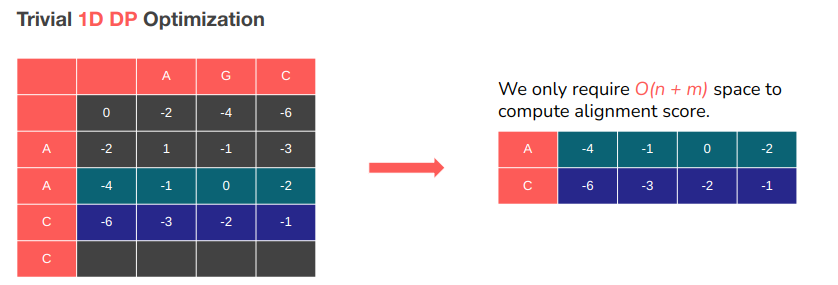
This optimizations provided a 100x speedup with no loss of accuracy, while only taking up O(n) space. The catch? We lose access to backtracking information. But in the future work we plan on combining the current work with the Hierschberg algorithm to restore access to this information while still having massive speedup.
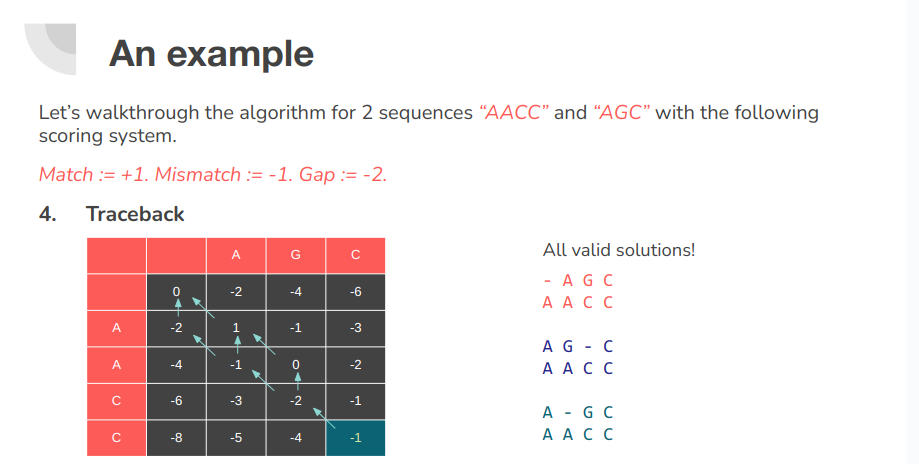
Pairwise sequence alignment is a way of arranging protein (or DNA) sequences to identify regions of similarity that may be a consequence of evolutionary relationships between the sequences. Encyclopedia of Bioinformatics and Computational Biology, 2019 There are two types of sequence alignment. Global and Local. Calculating a global alignment is a form of global optimization that "forces" the alignment to span the entire length of all query sequences. By contrast, local alignments identify regions of similarity within long sequences that are often widely divergent overall. Wikipedia - Sequence alignment
There are three primary components of alignment:
- Matches
- Mis-matches
- Gaps Each of these options are given some penalty or score. We then use these scores to compute the final score of alignment.
The Needleman-Wunsch algorithm uses dynamic programming (DP) to solve the global alignment problem optimally in O(nm) space and time complexity where n and m are the lengths of the two sequences we are trying to pairwise align.
Formally, let us define F(n, m) as a function that computes the global alignment score between two sequences A and B. This function can then be recursively defined as follows.
F(0, 0) = 0
F(i, 0) = F(i - 1, 0) - d
F(0, j) = F(0, j - 1) - d
F(i, j) = max { F(i - 1, j) - d,
F(i, j - 1) - d,
F(i - 1, j - 1) + s(A_i, B_i) }
Google Micro-Benchmark Library
One function run includes both reading files into memory and computing global alignment score.
Hardware: Ryzen 7 5800H (8c/16t @3.2GHz Base). 512KB / 4MB / 16MB Cache. 16GB RAM.
Test case information:
small/test_1/1.fasta → 10035bp small/test_1/2.fasta → 10200bp Order 1e4
small/test_2/1.fasta → 57474bp small/test_2/2.fasta → 56574bp Order 5e4
small/test_3/1.fasta → 162114bp small/test_3/2.fasta → 171823bp Order 1e5
small/test_4/1.fasta → 542868bp small/test_4/2.fasta → 536165bp Order 5e5
medium/1.fasta → 3147090bp medium/2.fasta → 3282708bp Order 3e6
- All benchmarks were run when CPU was idling at < 3% utilization across all cores as monitored by htop.
- Idling RAM usage was < 800MB.
- Caches were flushed before running the benchmarks.
- CPU Governor frequency was set to performance for more consistent results.
- Compiler (g++) flags:
-O3 -ffast-math -mavx2 -march=native -fopenmp
We’re often working with sequences that are of the magnitude 1e6 or even 1e9. Example: Human genome, size: 3,117,275,501 bp Wikipedia - Human Genome
1e6 ✕ 1e6 = 1e12 or ~1TB memory
1e6 ✕ 1e9 = 1e15 or ~1000TB memory
1e9 ✕ 1e9 = 1e18 or ~1 ExaByte memory!
Needleman-Wunsch requires O(nm) space to run… if we wish to also obtain the optimal alignment.
------------------------------------------------------------------------
Benchmark Time CPU Iterations
------------------------------------------------------------------------
needle2d_small_1_bench 167 ms 167 ms 4
needle2d_small_2_bench 4996 ms 4993 ms 1
needle1d_small_1_bench 86.9 ms 86.9 ms 8
needle1d_small_2_bench 2740 ms 2739 ms 1
needle1d_small_3_bench 23668 ms 23659 ms 1
needle1d_small_4_bench 257098 ms 256949 ms 1
- small/1 : 41.75ms / iter to 10.8625ms / iter (~4x speedup)
- small/2 : 4996ms to 2740ms (~2x speedup)
- small/3 : runs!
- small/4 : runs!
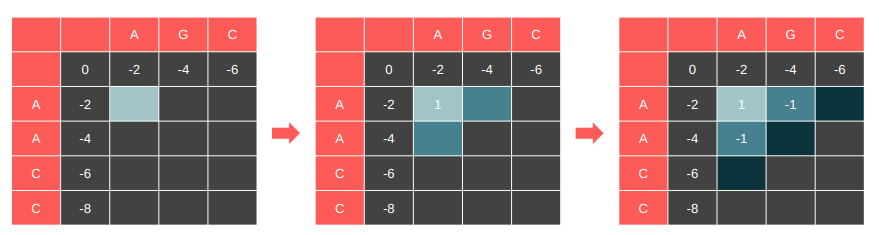
Dependence on (i - 1, j) means we cannot vectorize over columns.
Dependence on (i, j - 1) means we cannot vectorize over rows.
Also dependent on (i - 1, j - 1)
What to do?
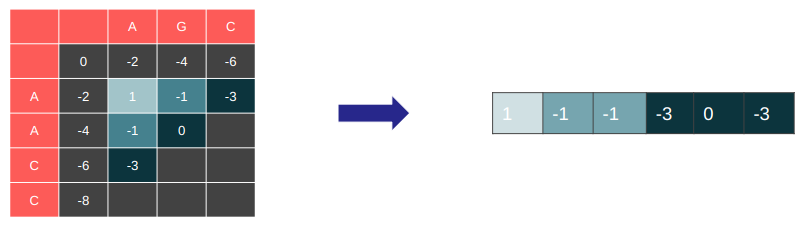
Let’s highlight the cells that have their dependencies satisfied and compute these cells in every iteration. This is essentially the dependency graph of our DP table.
It is parallelizable along the anti-diagonal.
The memory access pattern is terrible. Hard for pre-fetcher to learn pattern. Even if we can pre-fetch, terrible for spatial locality. Use blocking? Block the DP table into small chunks which individually fit in cache and compute the blocks along the anti-diagonal.
Cumbersome implementation. Will still have
Bcache misses per block for block sizeB. Cannot vectorize.
Restructure the DP table in memory so the wave-front is linearly stored in memory
Great memory access pattern. Good spatial locality.
Easy to implement 1D DP version with this structure → Good temporal locality.
__m128i _mm_cmpistrm (__m128i a, __m128i b, const int imm8)
Capable of performing bytewise character match and returning a mask. Slow instruction. Need to test.
------------------------------------------------------------------------
Benchmark Time CPU Iterations
------------------------------------------------------------------------
needle1d_small_1_bench 86.9 ms 86.9 ms 8
needle1d_small_2_bench 2740 ms 2739 ms 1
needle1d_small_3_bench 23668 ms 23659 ms 1
needle1d_small_4_bench 257098 ms 256949 ms 1
needle_diag_small_1_bench 14.7 ms 14.7 ms 48
needle_diag_small_2_bench 459 ms 459 ms 2
needle_diag_small_3_bench 4819 ms 4817 ms 1
needle_diag_small_4_bench 49874 ms 49848 ms 1
- small/1 : 10.8625ms / iter to 0.3062ms/iter (~35x speedup)
- small/2 : 2740ms to 229.5ms (~12x speedup)
- small/3 : 23668ms to 4819ms (~5x speedup)
- small/4 : 257098 ms to 49874ms (~5x speedup)
Each diagonal can be computed independently. For larger diagonals divide the diagonal into chunks and assign to each thread. Use parameter tuning to fine tune the decisions.
------------------------------------------------------------------------
Benchmark Time CPU Iterations
------------------------------------------------------------------------
needle_diag_small_1_bench 13.7 ms 13.7 ms 51
needle_diag_small_2_bench 432 ms 432 ms 2
needle_diag_small_3_bench 4676 ms 4673 ms 1
needle_diag_small_4_bench 49152 ms 49126 ms 1
needle_diag_par_small_1_bench 21.1 ms 21.1 ms 31
needle_diag_par_small_2_bench 319 ms 318 ms 2
needle_diag_par_small_3_bench 2318 ms 2317 ms 1
needle_diag_par_small_4_bench 23944 ms 23927 ms 1
needle_diag_par_medium_bench 3562798 ms 3548685 ms 1
- small/1 : 0.268ms / iter to 0.680ms/iter (Slower!)
- small/2 : 216ms to 159ms (~1.35x speedup)
- small/3 : 4676ms to 2318ms (~2x speedup)
- small/4 : 49152ms to 23944ms (~2x speedup)
- medium: runs! (~59.3 mins)
------------------------------------------------------------------------
Benchmark Time CPU Iterations
------------------------------------------------------------------------
needle2d_small_1_bench 167 ms 167 ms 4
needle2d_small_2_bench 4996 ms 4993 ms 1
needle1d_small_1_bench 86.9 ms 86.9 ms 8
needle1d_small_2_bench 2740 ms 2739 ms 1
needle1d_small_3_bench 23668 ms 23659 ms 1
needle_diag_small_1_bench 14.7 ms 14.7 ms 48 136x speedup!
needle_diag_small_2_bench 459 ms 459 ms 2
needle_diag_small_3_bench 4819 ms 4817 ms 1
needle_diag_small_4_bench 49874 ms 49848 ms 1
needle_diag_par_small_1_bench 23.7 ms 23.7 ms 30
needle_diag_par_small_2_bench 343 ms 343 ms 2 30x speedup :)
needle_diag_par_small_3_bench 2513 ms 2512 ms 1
needle_diag_par_small_4_bench 25883 ms 25816 ms 1
Hirschberg's algorithm is a clever modification of the Needleman–Wunsch Algorithm, which still takes O(nm) time, but needs only O(min{n, m}) space and is much faster in practice.
Wikipedia - Hirschberg's algorithm
However, while the time complexity is the same, it has a constant 2x computation requirement over Needleman-Wunsch.
Uses divide and conquer approach (Good for parallelization!)
Uses 1D Needleman-Wunsch in each recursion step. (We’ve already optimized that)
WIP. Just need to implement this :)