This repository is a coding exercise for the "Learning from Biosignals" course.
It is simplified version of the TinySleepNet, and its full implementation can be found in here.
- CUDA 12.1 (optional)
- mne 0.23.4
- matplotlib 3.3.4
- numpy 1.19.5
- pandas 1.1.5
- pyEDFlib 0.1.36
- torch 1.10.2
- scikit-learn 0.24.2
- scipy 1.5.4
- wget 3.2
First, you need to install miniconda to facilitate the creation of virtual Python environment. Please refer to this link for the installation.
Once you have installed the miniconda, you can run the following commands according to your operating system (OS).
For Windows/Ubuntu
conda create -n sleep python=3.6
conda activate sleep
pip install -r requirements.txt
conda install pytorch torchvision torchaudio pytorch-cuda=12.1 -c pytorch -c nvidiaFor MacOS
conda create -n sleep python=3.6
conda activate sleep
pip install -r requirements.txt
conda install pytorch::pytorch torchvision torchaudio -c pytorchIf you have problems when installing Pytorch, please refer to this link to find an alternative command to run.
python download_sleepedf.pypython prepare_sleepedf.pypython trainer.py --db sleepedf --gpu 0 --from_fold 0 --to_fold 19python predict.py --config_file config/sleepedf.py --model_dir out_sleepedf/train --output_dir out_sleepedf/predict --log_file out_sleepedf/predict.log
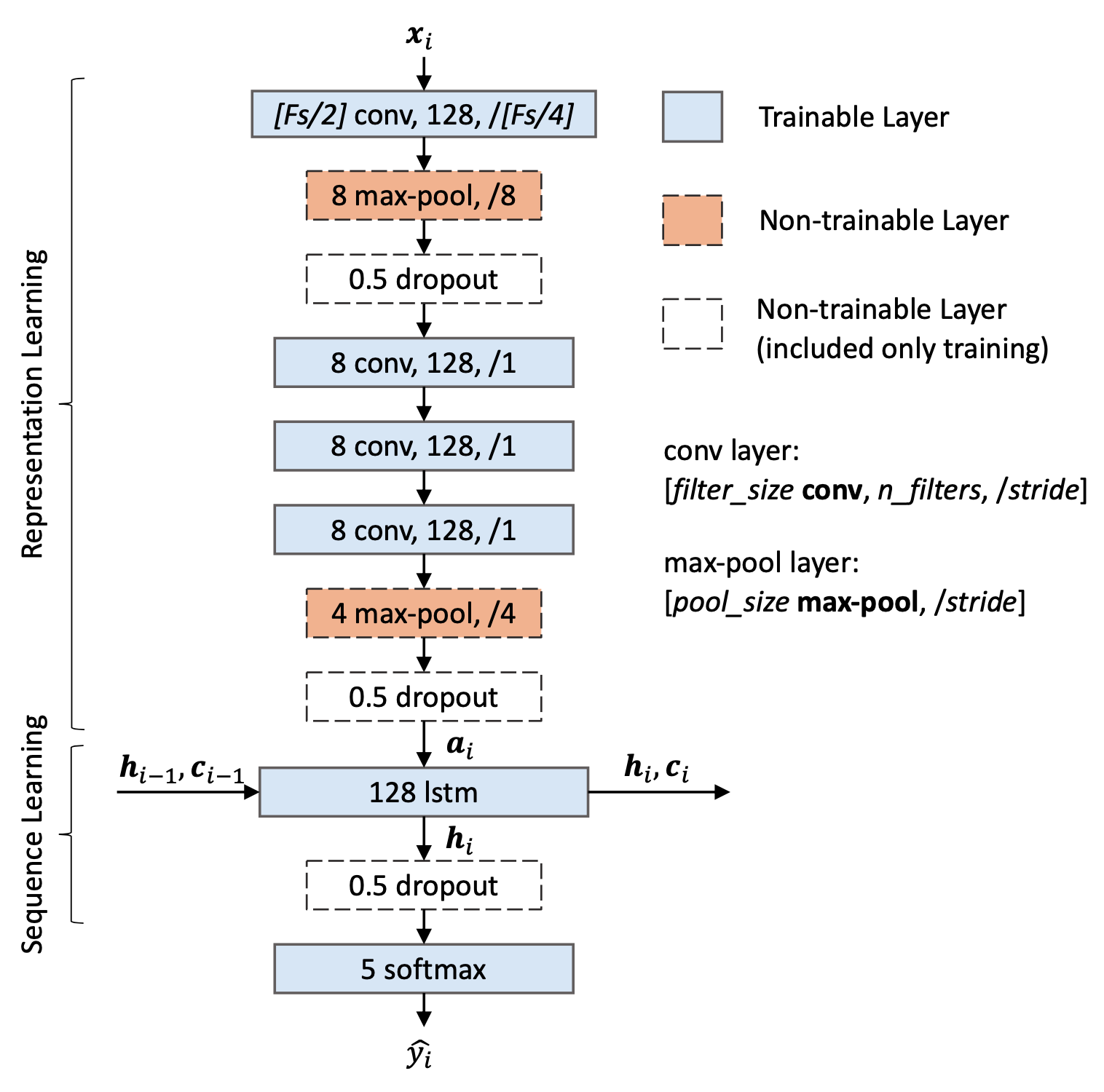
Modify the simple_model function in model.py to be the same as the Representation Learning part of the TinySleepNet model.
If you find this useful, please cite our work as follows:
@INPROCEEDINGS{Supratak2020,
title = {TinySleepNet: An Efficient Deep Learning Model for Sleep Stage Scoring based on Raw Single-Channel EEG},
author = {Supratak, Akara and Guo, Yike},
booktitle = {2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC),
year = {2020},
volume = {},
number = {},
pages = {641-644},
doi = {10.1109/EMBC44109.2020.9176741},
ISSN = {},
}