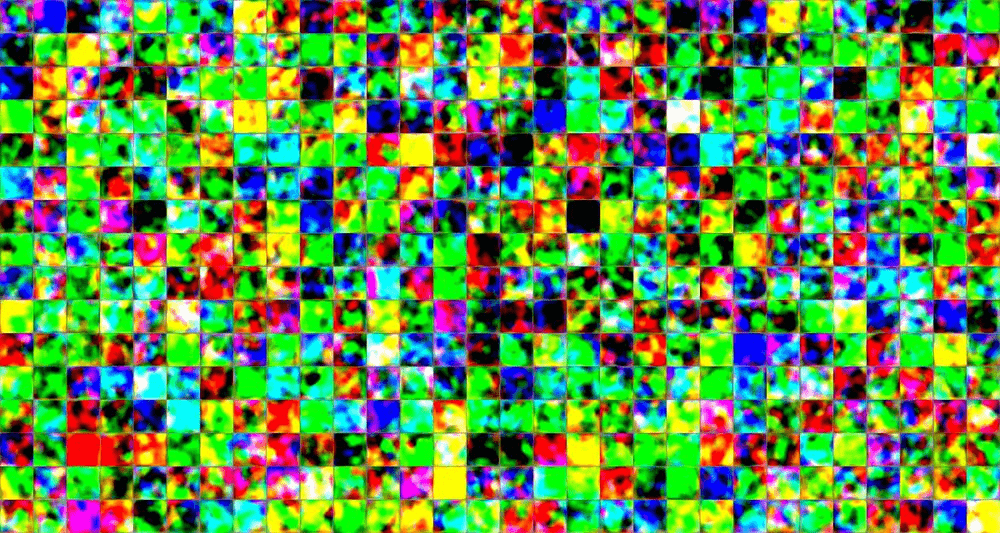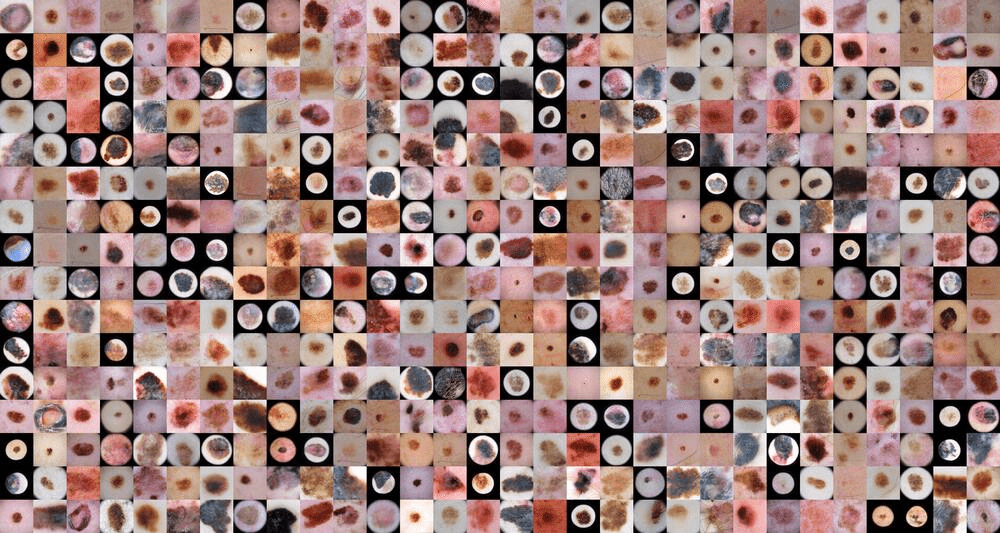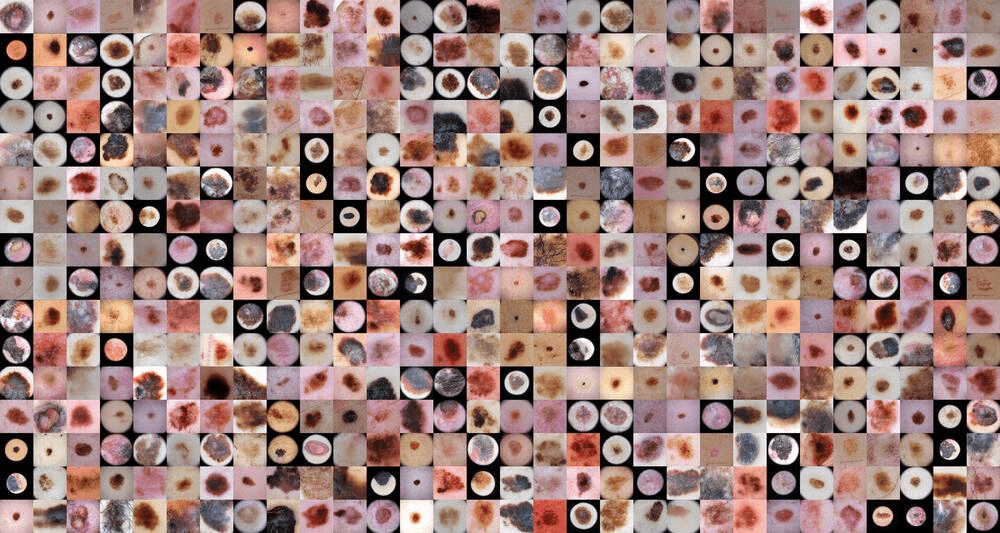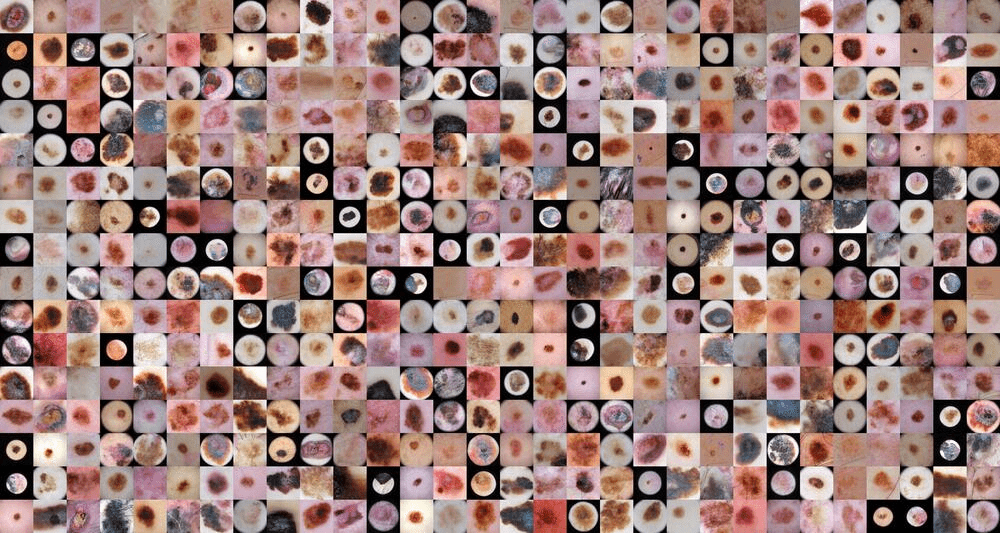
The usage of healthcare data in the development of artificial intelligence (AI) models is associated with issues around personal integrity and regulations. Patient data can usually not be freely shared and thus, the utility of it in creating AI solutions is limited. The main goal of the project was to explore GANs to generate synthetic data of skin lesions and test the performance of DL models train on that type of data in comparison to trained only on real one.
- Linux and Windows are supported, but we recommend Linux for performance and compatibility reasons.
- 1–8 high-end NVIDIA GPUs with at least 12 GB of memory. We have done all testing and development using NVIDIA DGX-1 with 8 Tesla V100 GPUs.
- 64-bit Python 3.7 and PyTorch 1.7.1. See https://pytorch.org/ for PyTorch install instructions.
- CUDA toolkit 11.0 or later. Use at least version 11.1 if running on RTX 3090. (Why is a separate CUDA toolkit installation required? See comments in #2.)
- Python libraries:
pip install click requests tqdm pyspng ninja imageio-ffmpeg==0.4.3. We use the Anaconda3 2020.11 distribution which installs most of these by default. - Docker users: use the provided Dockerfile to build an image with the required library dependencies.
The code relies heavily on custom PyTorch extensions that are compiled on the fly using NVCC. On Windows, the compilation requires Microsoft Visual Studio. We recommend installing Visual Studio Community Edition and adding it into PATH using "C:\Program Files (x86)\Microsoft Visual Studio\<VERSION>\Community\VC\Auxiliary\Build\vcvars64.bat".
Pre-trained networks are stored as *.pkl files that can be referenced using local filenames or URLs:
# Generate curated MetFaces images without truncation (Fig.10 left)
python generate.py --outdir=out --trunc=1 --seeds=85,265,297,849 \
--network=https://nvlabs-fi-cdn.nvidia.com/stylegan2-ada-pytorch/pretrained/metfaces.pkl
# Generate uncurated MetFaces images with truncation (Fig.12 upper left)
python generate.py --outdir=out --trunc=0.7 --seeds=600-605 \
--network=https://nvlabs-fi-cdn.nvidia.com/stylegan2-ada-pytorch/pretrained/metfaces.pkl
# Generate class conditional CIFAR-10 images (Fig.17 left, Car)
python generate.py --outdir=out --seeds=0-35 --class=1 \
--network=https://nvlabs-fi-cdn.nvidia.com/stylegan2-ada-pytorch/pretrained/cifar10.pkl
# Style mixing example
python style_mixing.py --outdir=out --rows=85,100,75,458,1500 --cols=55,821,1789,293 \
--network=https://nvlabs-fi-cdn.nvidia.com/stylegan2-ada-pytorch/pretrained/metfaces.pklOutputs from the above commands are placed under out/*.png, controlled by --outdir. Downloaded network pickles are cached under $HOME/.cache/dnnlib, which can be overridden by setting the DNNLIB_CACHE_DIR environment variable. The default PyTorch extension build directory is $HOME/.cache/torch_extensions, which can be overridden by setting TORCH_EXTENSIONS_DIR.
Docker: You can run the above curated image example using Docker as follows:
docker build --tag sg2ada:latest .
./docker_run.sh python3 generate.py --outdir=out --trunc=1 --seeds=85,265,297,849 \
--network=https://nvlabs-fi-cdn.nvidia.com/stylegan2-ada-pytorch/pretrained/metfaces.pklNote: See docker_run.sh for more information.
Datasets are stored as uncompressed ZIP archives containing uncompressed PNG files and a metadata file dataset.json for labels.
Custom datasets can be created from a folder containing images; see python dataset_tool.py --help for more information. Alternatively, the folder can also be used directly as a dataset, without running it through dataset_tool.py first, but doing so may lead to suboptimal performance.
ISIC 2020: Download the ISIC 2020 dataset and create ZIP archive:
python dataset_tool.py --source=/tmp/isic-dataset --dest=~/datasets/isic256x256.zip --width=256 --height=256In its most basic form, training new networks boils down to:
python train.py --outdir=~/training-runs --data=~/mydataset.zip --gpus=1 --dry-run
python train.py --outdir=~/training-runs --data=~/mydataset.zip --gpus=1The first command is optional; it validates the arguments, prints out the training configuration, and exits. The second command kicks off the actual training.
In this example, the results are saved to a newly created directory ~/training-runs/<ID>-mydataset-auto1, controlled by --outdir. The training exports network pickles (network-snapshot-<INT>.pkl) and example images (fakes<INT>.png) at regular intervals (controlled by --snap). For each pickle, it also evaluates FID (controlled by --metrics) and logs the resulting scores in metric-fid50k_full.jsonl (as well as TFEvents if TensorBoard is installed or loged to wandb if one has an account).
The training configuration can be further customized with additional command line options:
--aug=noaugdisables ADA.--cond=1enables class-conditional training (requires a dataset with labels).--mirror=1amplifies the dataset with x-flips. Often beneficial, even with ADA.--resume=ffhq1024 --snap=10performs transfer learning from FFHQ trained at 1024x1024.--resume=~/training-runs/<NAME>/network-snapshot-<INT>.pklresumes a previous training run.--gamma=10overrides R1 gamma. We recommend trying a couple of different values for each new dataset.--aug=ada --target=0.7adjusts ADA target value (default: 0.6).--augpipe=blitenables pixel blitting but disables all other augmentations.--augpipe=bgcfncenables all available augmentations (blit, geom, color, filter, noise, cutout).
Please refer to python train.py --help for the full list.
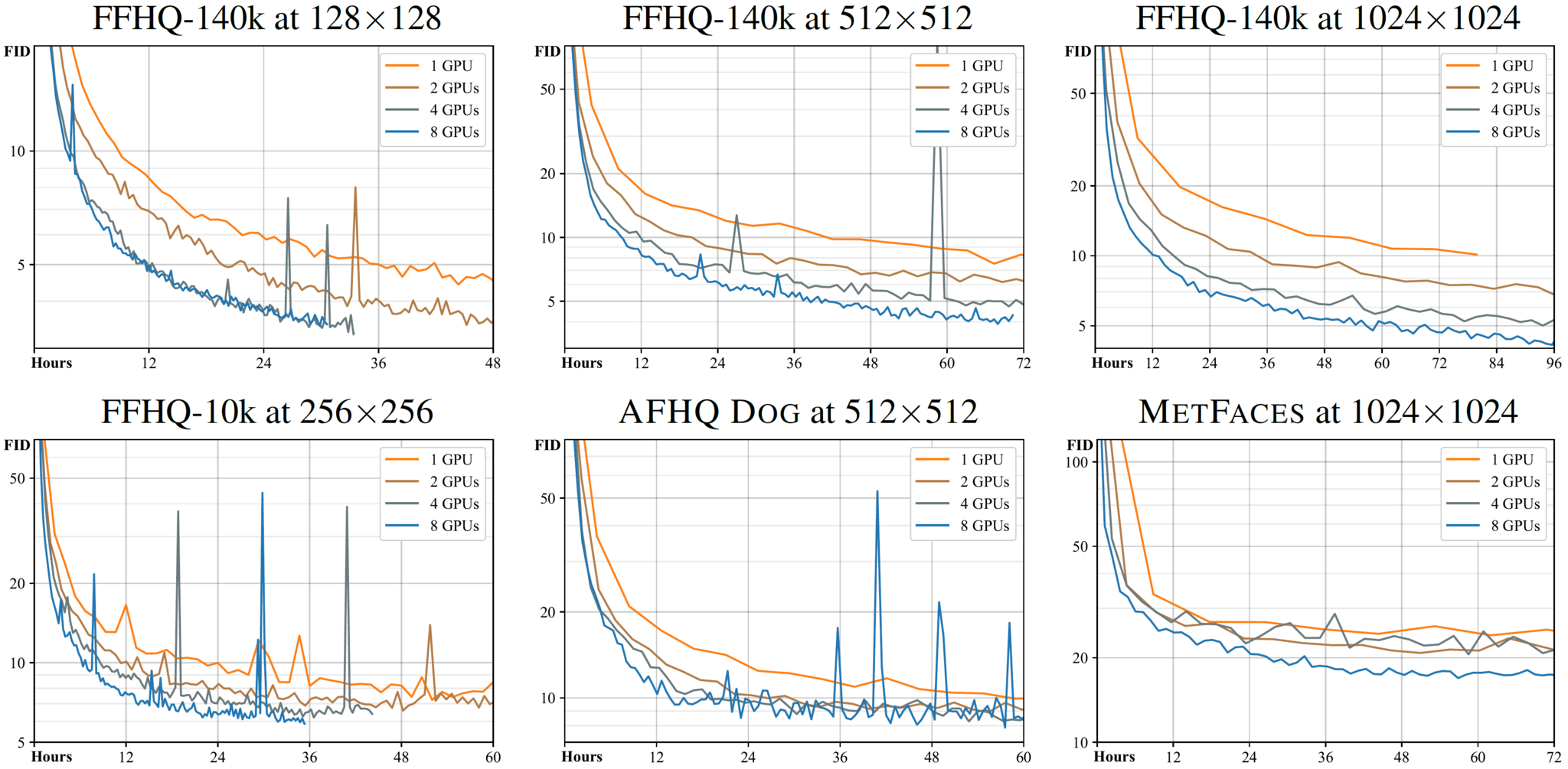
The total training time depends heavily on resolution, number of GPUs, dataset, desired quality, and hyperparameters. The following table lists expected wallclock times to reach different points in the training, measured in thousands of real images shown to the discriminator ("kimg"):
| Resolution | GPUs | 1000 kimg | 25000 kimg | sec/kimg | GPU mem | CPU mem |
|---|---|---|---|---|---|---|
| 128x128 | 1 | 4h 05m | 4d 06h | 12.8–13.7 | 7.2 GB | 3.9 GB |
| 128x128 | 2 | 2h 06m | 2d 04h | 6.5–6.8 | 7.4 GB | 7.9 GB |
| 128x128 | 4 | 1h 20m | 1d 09h | 4.1–4.6 | 4.2 GB | 16.3 GB |
| 128x128 | 8 | 1h 13m | 1d 06h | 3.9–4.9 | 2.6 GB | 31.9 GB |
| 256x256 | 1 | 6h 36m | 6d 21h | 21.6–24.2 | 5.0 GB | 4.5 GB |
| 256x256 | 2 | 3h 27m | 3d 14h | 11.2–11.8 | 5.2 GB | 9.0 GB |
| 256x256 | 4 | 1h 45m | 1d 20h | 5.6–5.9 | 5.2 GB | 17.8 GB |
| 256x256 | 8 | 1h 24m | 1d 11h | 4.4–5.5 | 3.2 GB | 34.7 GB |
| 512x512 | 1 | 21h 03m | 21d 22h | 72.5–74.9 | 7.6 GB | 5.0 GB |
| 512x512 | 2 | 10h 59m | 11d 10h | 37.7–40.0 | 7.8 GB | 9.8 GB |
| 512x512 | 4 | 5h 29m | 5d 17h | 18.7–19.1 | 7.9 GB | 17.7 GB |
| 512x512 | 8 | 2h 48m | 2d 22h | 9.5–9.7 | 7.8 GB | 38.2 GB |
| 1024x1024 | 1 | 1d 20h | 46d 03h | 154.3–161.6 | 8.1 GB | 5.3 GB |
| 1024x1024 | 2 | 23h 09m | 24d 02h | 80.6–86.2 | 8.6 GB | 11.9 GB |
| 1024x1024 | 4 | 11h 36m | 12d 02h | 40.1–40.8 | 8.4 GB | 21.9 GB |
| 1024x1024 | 8 | 5h 54m | 6d 03h | 20.2–20.6 | 8.3 GB | 44.7 GB |
The above measurements were done using NVIDIA Tesla V100 GPUs with default settings (--cfg=auto --aug=ada --metrics=fid50k_full). "sec/kimg" shows the expected range of variation in raw training performance, as reported in log.txt. "GPU mem" and "CPU mem" show the highest observed memory consumption, excluding the peak at the beginning caused by torch.backends.cudnn.benchmark.
In typical cases, 25000 kimg or more is needed to reach convergence, but the results are already quite reasonable around 5000 kimg. 1000 kimg is often enough for transfer learning, which tends to converge significantly faster. The following figure shows example convergence curves for different datasets as a function of wallclock time, using the same settings as above:
Note: --cfg=auto serves as a reasonable first guess for the hyperparameters but it does not necessarily lead to optimal results for a given dataset. We recommend trying out at least a few different values of --gamma for each new dataset.
By default, train.py automatically computes FID for each network pickle exported during training. We recommend inspecting metric-fid50k_full.jsonl (or TensorBoard) at regular intervals to monitor the training progress. When desired, the automatic computation can be disabled with --metrics=none to speed up the training slightly (3%–9%).
Additional quality metrics can also be computed after the training:
# Previous training run: look up options automatically, save result to JSONL file.
python calc_metrics.py --metrics="fid50k_full,kid50k_full,pr50k3_full,ppl2_wend" \
--mirror=1 --data=~/datasets/isic256x256.zip --network=~/network-snapshot-000000.pklTo find the matching latent vector for a given image file, run:
python projector.py --outdir=out --target=~/mytargetimg.png \
--class_label=1 --network=~/pretrained/conditionalGAN.pklThe above command saves the projection target out/target.png, result out/proj.png, latent vector out/projected_w.npz, and progression video out/proj.mp4. You can render the resulting latent vector by specifying --projected_w for generate.py for specific melanoma class:
python generate.py --outdir=out --projected_w=out/projected_w.npz \
--class=1 --network=~/pretrained/conditionalGAN.pklIn our studies generated synthetic images were used in binary classification task between melanoma and non-melanoma cases. To run training with Efficientnet-B2 use following command:
python melanoma_classifier.py --syn_data_path=~/generated/ \
--real_data_path=~/melanoma-external-malignant-256/ \
--synt_n_imgs="0,15"In above example --syn_data_path argument indicates path for synthetic images,
--real_data_path - real images and --synt_n_imgs stands for n non-melanoma, k melanoma synthetic images (measured in kimg) to add to the real data. We reported our studis using wandb (use --wandb_flag argument to report accuracy and loss for your own experiments). --only_reals flag enable training only for real images, while --only_syn will allow to take all artificial images from directory with synthetic images.
To make a diagnosis using trained model use predict.py script.
embeddings_projector.py performs the two following tasks:
-
Project embeddings of a CNN used as feature extractor. (
--use_cnn) -
Project w-vectors.
This generates a metadata.tsv, tensors.tsv and (optionally using --sprite flag) a sprite of the images. These files can be uploaded in the Tensorboard Projector , which graphically represent these embeddings.
We additionaly calculated cosine distances between the CNN embeddings from the tsv file.
For details see read_tsv.py.
python ./CNN_embeddings_projector/read_tsv.py --metadata=metadata.tsv \
--embeddings_path=tensors.tsv --save_path=distances.txtOur results are available in form of papers:
Assessing GAN-Based Generative Modeling on Skin Lesions Images
@InProceedings{10.1007/978-3-031-37649-8_10,
author="Carrasco Limeros, Sandra
and Majchrowska, Sylwia
and Zoubi, Mohamad Khir
and Ros{\'e}n, Anna
and Suvilehto, Juulia
and Sj{\"o}blom, Lisa
and Kjellberg, Magnus",
editor="Biele, Cezary
and Kacprzyk, Janusz
and Kope{\'{c}}, Wies{\l}aw
and Owsi{\'{n}}ski, Jan W.
and Romanowski, Andrzej
and Sikorski, Marcin",
title="Assessing GAN-Based Generative Modeling on Skin Lesions Images",
booktitle="Digital Interaction and Machine Intelligence",
year="2023",
publisher="Springer Nature Switzerland",
address="Cham",
pages="93--102",
isbn="978-3-031-37649-8"
}
GAN-based generative modelling for dermatological applications -- comparative study
@misc{https://doi.org/10.48550/arxiv.2208.11702,
doi = {10.48550/ARXIV.2208.11702},
url = {https://arxiv.org/abs/2208.11702},
author = {Carrasco Limeros, Sandra and Majchrowska, Sylwia and Zoubi, Mohamad Khir and Rosén, Anna and Suvilehto, Juulia and Sjöblom, Lisa and Kjellberg, Magnus},
title = {GAN-based generative modelling for dermatological applications -- comparative study},
publisher = {arXiv},
year = {2022},
}
The (de)biasing effect of GAN-based augmentation methods on skin lesion images
@inproceedings{Mikolajczyk:22,
author="Miko{\l}ajczyk, Agnieszka and Majchrowska, Sylwia and Carrasco Limeros, Sandra",
editor="Wang, Linwei and Dou, Qi and Fletcher, P. Thomas and Speidel, Stefanie and Li, Shuo",
title="The (de)biasing Effect of GAN-Based Augmentation Methods on Skin Lesion Images",
booktitle="Medical Image Computing and Computer Assisted Intervention -- MICCAI 2022",
year="2022",
publisher="Springer Nature Switzerland",
address="Cham",
pages="437--447",
isbn="978-3-031-16452-1"
}
This repository depends on NVIDIA Corporation's repository. Copyright © 2021, NVIDIA Corporation. All rights reserved.
This work is made available under the Nvidia Source Code License.
The project was developed during the first rotation of the Eye for AI Program at the AI Competence Center of Sahlgrenska University Hospital. Eye for AI initiative is a global program focused on bringing more international talents into the Swedish AI landscape.