The Polar pipeline and all its dependencies are Linux based, typically running under Linux
operating system, preferably (but not necessarily) on a computer cluster. The included test
set can run on a laptop in under 5 minutes. There are several options for installation, detailed below.
git clone https://github.com/peradastra/PAP.gitor...
curl -sSL -o PAP.zip https://github.com/peradastra/PAP/master.zip && \
mkdir -p PAP && \
unzip PAP.zip -d PAPIf you already have a Anaconda/Miniconda installation then you can create a conda environment using the provided
environment definition.
- Create the conda environment
conda env create -n pap_conda_env -f PAP/pap_conda_env.yml- Activate PAP conda environment
conda activate pap_conda_envcd PAP && \
bash run_pap.sh -d Library001Usage: run_pap.sh [-d TOP_DIR] [-t THREADS] -h
-d Top level directory which must contain a subdirectory (fastq/) with fastq files
-t Number of threads for BWA alignment (Default: 16)
-h Print this help and exit
Place the paired-end sequenced reads in a folder labeled fastq. For example, if your experiment is called "Library001", you should have a folder labeled "Library001," and it should contain one subfolder labeled "fastq" with the fastq files in it. The fastqs can be zipped or unzipped, and there can be multiple pairs. This directory structure is shown below in the form of a tree structure.
Library001
└── fastq
├── library001_R1.fastq.gz
└── library001_R2.fastq.gz
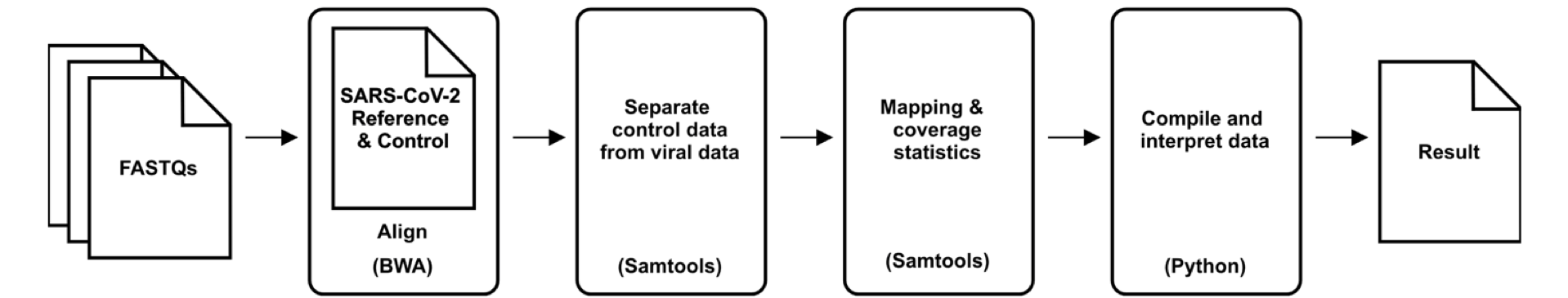
The pipeline will create an "aligned" folder, an "debug" folder and a "final" folder under "Library001." The "alignments" folder will contain alignments. The "debug" folder will contain the error and log files. The "final" folder will contain the final results of the pipeline.
Library001
├── fastq
│ ├── library001_R1.fastq.gz
│ └── library001_R2.fastq.gz
└── pap
├── aligned
│ ├── all_alignment_stats.txt
│ ├── classified_alignments.bam
│ └── viral_alignment_stats.txt
├── debug
│ ├── align.out
│ ├── all_stats.out
│ ├── dedup.out
* *
* *
* *
│ ├── matefix.out
│ ├── recombo.out
│ ├── sort.out
│ └── viral_stats.out
└── final
├── qc_stats.txt
└── result.csv