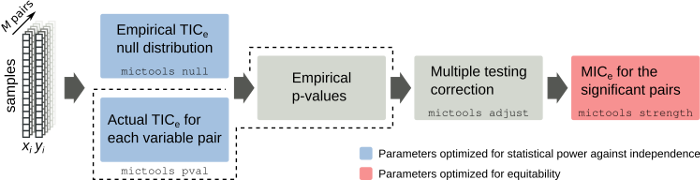
MICtools is an open source pipeline which combines the TIC_e and MIC_e measures [Reshef2016] into a two-step procedure that allows to identify relationships of various degrees of complexity in large datasets. TIC_e is used to perform an efficient high throughput screening of all the possible pairwise relationships and a permutation based appraoch is used to assess their significance. MIC_e is then used to rank the subset of significant associations on the bases of their strength.
The MICtools pipeline can be broken into 4 steps (see the figure above):
- given M variables pairs x_i and y_i measured in n samples, the empirical TIC_e null distribution is estimated by permutation;
- TIC_e statistics and the associated empirical p-values are computed for all variable pairs;
- p-values are corrected for multiplicity in order to control the family-wise error rate (FWER) or the false discovery rate (FDR);
- finally, the strengths of the relationships called significant are estimated using the MIC_e estimator.
Table of contents
- local
We suggest to install the Python and the GCC compiler through the package manager (In Mac OS X, we recommend to install them using Homebrew) (e.g. on Ubuntu/Debian):
Then, upgrade pip and install setuptools:
Finally, install mictools:
- Install Docker for Linux, Mac OS X or Windows.
- Run the
Docker Quickstart Terminal(Mac OS X, Windows) or thedockerdaemon (Linux,sudo service docker start). - Follow the instructions at https://hub.docker.com/r/minepy/mictools/.
If you are installing from source, the following dependences must be installed: Python >= 2.7, Click >= 5.1, numpy >= 1.7.0, scipy >= 0.13, pandas >= 0.17.0, matplotlib >= 1.2.0,<2, statsmodels >= 0.6.1, minepy >= 1.2. We suggest to install these dependences using the OS package manager (Linux), Homebrew (macOS/OS X) or pip.
Download the latest stable version from https://github.com/minepy/mictools/releases and complete the installation:
MICtools can be used to investigate variable associations in different types of experimental scenarios:
- single dataset X, with M variables and N samples: to evaluate the M+(M-1)/2 possible associations;
- two datasets, X (MxN) and Y (KxN) (parameter -y/--yvars): to evaluate all the pairwise relationships between the variables of the two datasets (for a total of MxK associations). Note that the number samples (N) in the datasets X and Y must be the same.
- * two datasets, X (MxN) and Y (KxN): to evaluate all the rowwise
relationships (see -r/--rowwise), i.e. only the variables pairs X_i and Y_i (for each i in min(M, K)) will be tested;
In all the abovementioned cases the analysis will be performed within each class independently if the sample classes are provided (see -l/--labels and -t/--target).
MICtools is implemented as a single command (mictools'') with the following subcommands:nullCompute the TIC_e null distribution.mergenullMerge multiple TIC_e null distributions.pvalCompute TIC_e p-values.adjustMultiple testing correction.strengthCompute the strength (MIC_e). Runmictools SUBCOMMAND --helpfor the documentation of each specific step. Tutorial -------- We analyze the "Datasaurus" synthetic dataset generated following the approach discussed at https://www.autodeskresearch.com/publications/samestats ([Matejka2017]_). The dataset contains 26 variables linked by 13 relationships which have the same summary statistics (e.g. the Pearson's correlation), but are very different in appearance. The dataset was modified in order to destroy secondary associations. In this example we test the entire set of possible associations (for a total of 26*(26-1)/2 = 325 relationships). Preparation ^^^^^^^^^^^ Go to theexamplesfolder: .. code-block:: sh cd examples Select the Datasaurus dataset and the output folder: .. code-block:: sh X=datasaurus.txt ODIR=datasaurus_results mkdir $ODIR Empirical TIC_e null distribution ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ Compute the empirical TIC_e null distribution (with 200,000 permutations, default value): .. code-block:: sh mictools null $X $ODIR/null_dist.txt The output filenull_dist.txtis a TAB-delimited file which contains the null distrubution: ===== ======== ======== ========= ============ Class BinStart BinEnd NullCount NullCountCum ===== ======== ======== ========= ============ None 0.000000 0.000100 0 200000 None 0.000100 0.000200 0 200000 None 0.000200 0.000300 0 200000 ... ... ... ... ... ===== ======== ======== ========= ============ The first column (Class) contains the class membership (in this particular case no sample classes were provided),BinStartandBinEnddefine the TIC_e range andNullCountandNullCountCumare distribution and the cumulative distribution, respectively. TIC_e p-values ^^^^^^^^^^^^^^ Compute the TIC_e statistics and the associated empirical p-values for all variable pairs: .. code-block:: sh mictools pval $X $ODIR/null_dist.txt $ODIR The command will return in the output directory the following:obs_dist.txtthe observed TICe distribution in the same format ofnull_dist.txtobs.txtTAB-delimited file containing the observed TICe values for each variable pair tested: ====== ========== ======== Var1 Var2 None ====== ========== ======== away_x bullseye_x 0.029476 away_x circle_x 0.018211 away_x dino_x 0.050720 ... ... ... ====== ========== ========pval.txtTAB-delimited file containing the empirical p-values for each variable pairpval_None.pngthe p values distribution plot: .. image:: docs/images/pval_None.png Multiple testing correction ^^^^^^^^^^^^^^^^^^^^^^^^^^^ Correct the p-values for multiplicity in order to control the false discovery rate (FDR, default method); .. code-block:: sh mictools adjust $ODIR/pval.txt $ODIR The command returns in the OUTPUT directory the following files:pval_adj.txtadjusted p values for each variable pair tested, in the same format ofpval.txtpi0_None.pngsince the correction method is the Storey's qvalue, the command returns a plot with the estimated pi_0 versus the tuning parameter lambda: .. image:: docs/images/pi0_None.png Strength of significant associations ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ Finally, the strengths of the relationships called significant are estimated using MIC_e. By default the significance level is set to 0.05: .. code-block:: sh mictools strength $X $ODIR/pval_adj.txt $ODIR/strength.txt The output filestrength.txt`` is a TAB-delimited file, containing for each significant association the (corrected) TIC_e p-values, the Pearson's correlations, the Spearman's coefficients and finally the strengths, i.e. the MIC_e values:
| Class | Var1 | Var2 | TICePVal | PearsonR | SpearmanRho | MICe |
|---|---|---|---|---|---|---|
| None | bullseye_x | bullseye_y | 3.833704e-02 | -0.068586 | -0.078734 | 0.424553 |
| None | circle_x | circle_y | 4.723013e-04 | -0.068343 | -0.077292 | 0.631458 |
| None | dots_x | dots_y | 1.983666e-02 | -0.060342 | -0.126174 | 0.500185 |
| None | slant_up_x | slant_up_y | 1.593666e-02 | -0.068609 | -0.086098 | 0.355019 |
| None | star_x | star_y | 4.723013e-04 | -0.062961 | -0.051445 | 0.633117 |
| None | x_shape_x | x_shape_y | 4.723013e-04 | -0.065583 | -0.020535 | 0.566703 |
- Matejka2017
J. Matejka and G. Fitzmaurice. Same Stats, Different Graphs: Generating Datasets with Varied Appearance and Identical Statistics through Simulated Annealing. ACM SIGCHI Conference on Human Factors in Computing Systems, 2017.
- Reshef2016
Yakir A. Reshef, David N. Reshef, Hilary K. Finucane and Pardis C. Sabeti and Michael Mitzenmacher. Measuring Dependence Powerfully and Equitably. Journal of Machine Learning Research, 2016.