The epiLION project is aimed to provide a unified identifier for major lipids, especially oxidized lipids in the epilipidome.
-
Main Features
- Optimized for manual interpretation and computer processing
- Unified modification controlled vocabularies
- Unified position specific annotations
- Special optimization for prostane containing lipids
- Suitable for head group modified phospholipids
- Hierarchical abbreviation system
- Capable to parse fuzzy site unspecific annotations
-
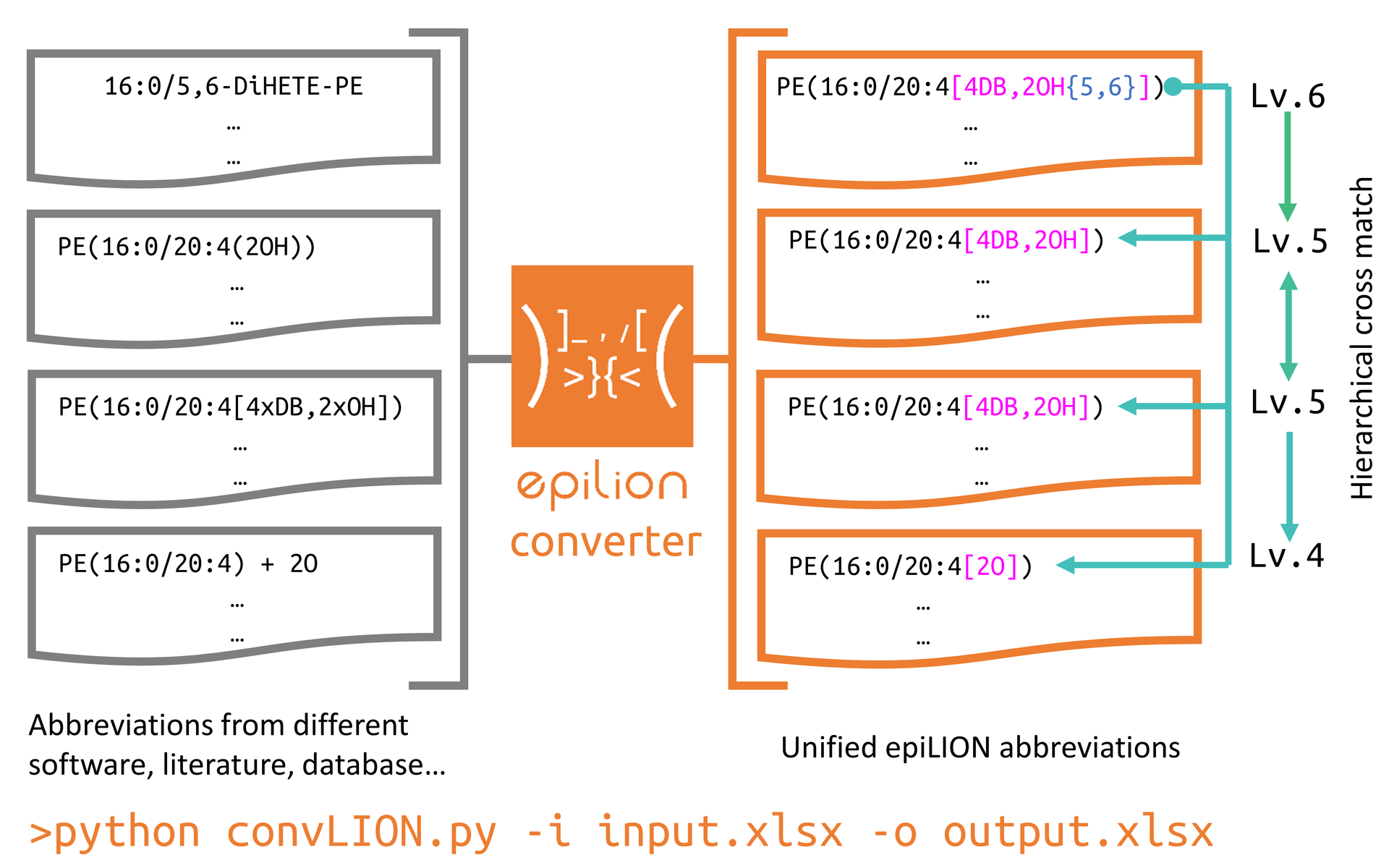
epiLION abbreviation converter
- Convert different abbreviations to uniformed epiLION abbreviations
-
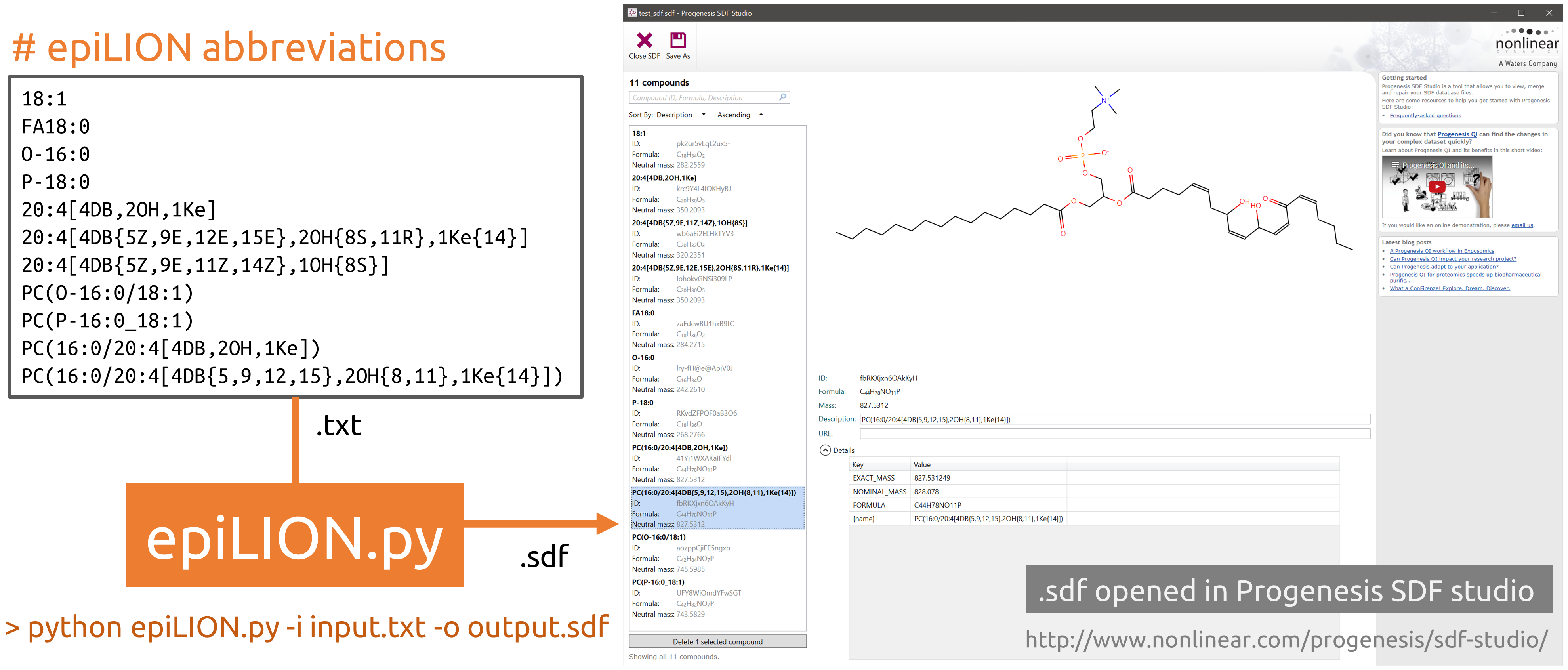
epiLION structure and MS property generator
- Abbreviation to SMILES/ MOL / SDF conversion using python
-
Currently supported modifications
DB: C=C bondOH: hydroxyHp: hydroperoxyNH2: aminoMe: methylKe: keto/oxoEp: epoxySH: thioMy: methyleneBr: bromoCl: chloroF: fluoroCN: cyano
-
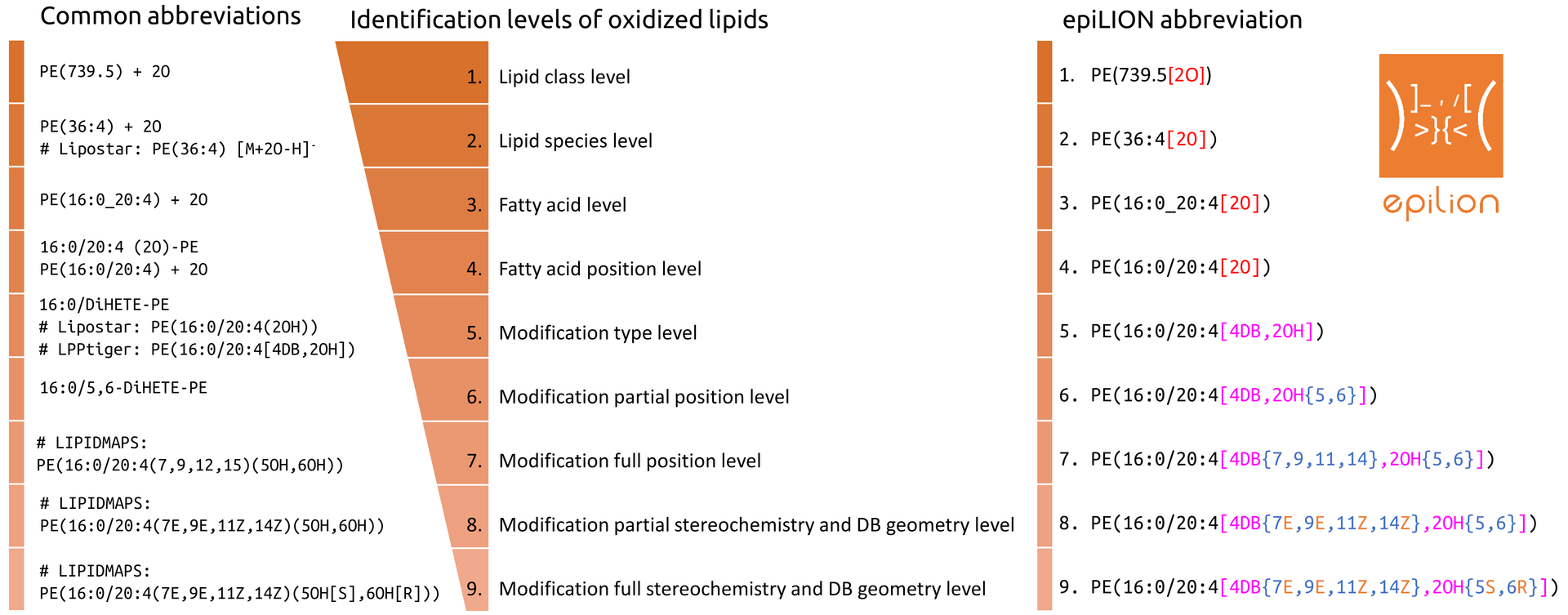
Example of epiLION abbreviations
-
Fatty acids
- FA18:0
- O-16:0
- P-18:0
- 20:4[4DB,2OH,1Ke]
- 20:4[4DB{5Z,9E,11Z,14Z},1OH{8S}]
- 20:4[4DB{5Z,9E,12E,15E},2OH{8S,11R},1Ke{14}]
-
Phospholipids
- PC(O-16:0/18:1)
- PC(P-16:0_18:1)
- PC(16:0/20:4[4DB,2OH,1Ke])
- PC(16:0/20:4[4DB{5,9,12,15},2OH{8,11},1Ke{14}])
-
-
Download the source code as zip file for your system
-
Download epiLION source Code as .zip. Please notice the date and version of LipidHunter source code.
-
Professional users can use
gitto clone the whole repository, please make sure that you switched to the correct branch. -
Only the released version is recommended for real data processing. Other development branches may lead to unknown issues and miss interpretation of the data.
-
-
Rename the downloaded file to
epiLION.zip -
Unzip
epiLION.zipfile to any folder. -
Install epiLION source code
-
epiLION is developed under python 3.6+.
-
The best way is to use virtual environment such as
conda -
Main dependencies are:
-
Data processing:
pandas -
SDF generation:
rdkit
-
-
Test source code installation
-
epiLION is configured to use travis-ci with
py.testto test on Windows, Linux, and macOS. -
you can also use py.test to test epiLION in your python environment
-
unit test for epiLION converter is provided in
Test/test_convLION.py -
unit test for epiLION generator is provided in
Test/test_epiLION.py
-
-
-
-
Run epiLION converter
-
Run
python convLION.py -i Test/TestInput/test_crosscheck.xlsx -o Test/TestOutput/test_crosscheck_output.xlsx -
convLION read list of different abbreviations in the
test_crosscheck.xlsxand generate the converted epiLION abbreviations in the output file. -
convLION can read an write both
.xlsxand.csvfile. -
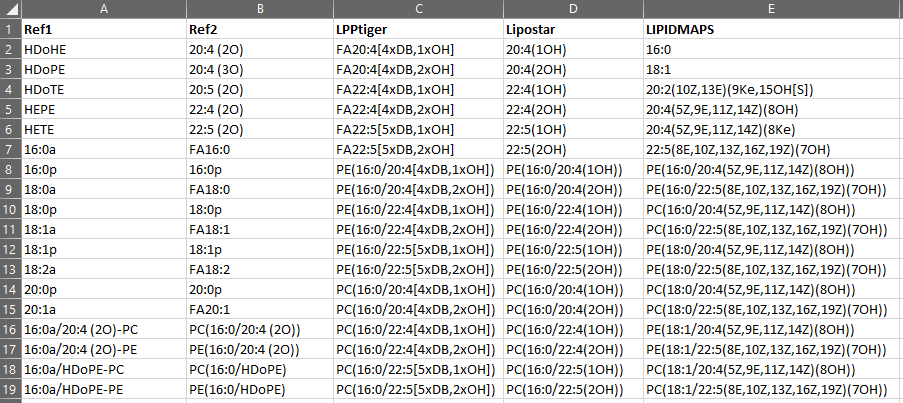
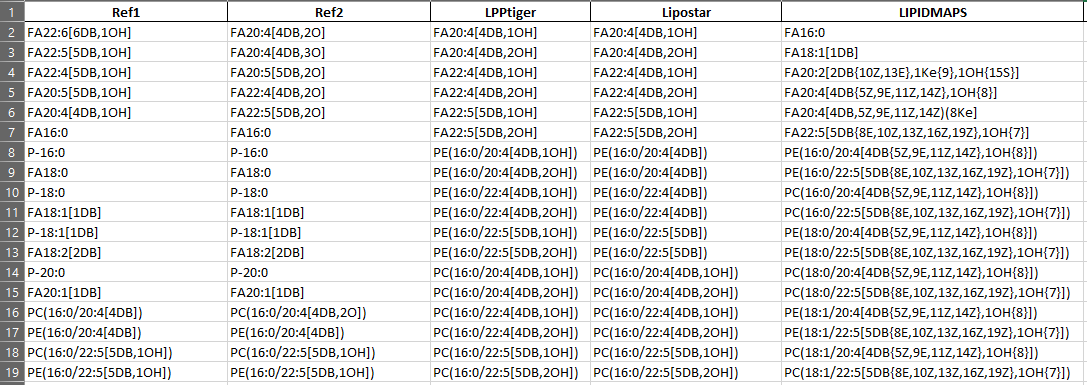
sample input
-
sample output
-
-
Run epiLION generator
-
Run
python epiLION.py -i Test/TestInput/test_names.txt -o Test/TestOutput/test_sdf.sdf -
epiLION read list of epiLION abbreviations in the
test_names.txtand generate the structure in a combined sdf file.
-
-
SDF output
-
The sdf file is generated by using
rdkithttps://www.rdkit.org -
The sdf output can be used by:
-
Progenesis SDF studio, free for academic use: http://www.nonlinear.com/progenesis/sdf-studio/
-
ChemAxon Instant Jchem, academic license available: https://chemaxon.com/products/instant-jchem
-
-
-
Errors/bugs
In case you experienced any problems with running LipidHunter
please report an issue in the issue tracker or contact us.
-
LipidHunter is Dual-licensed
-
For academic and non-commercial use:
GPLv2 License: -
For commercial use: please contact the develop team by email.
-
-
Please cite our publication in an appropriate form.
- Ni, Zhixu, Laura Goracci, Gabriele Cruciani, and Maria Fedorova. "Computational solutions in redox lipidomics–Current strategies and future perspectives." Free Radical Biology and Medicine (2019). - DOI: 10.1016/j.freeradbiomed.2019.04.027
- Report any issues here: https://github.com/SysMedOs/epiLION/issues
We acknowledge all projects that supports the development of LipidHunter:
-
BMBF - Federal Ministry of Education and Research Germany:
-
e:Med Systems Medicine Network:
-
SysMedOS Project :