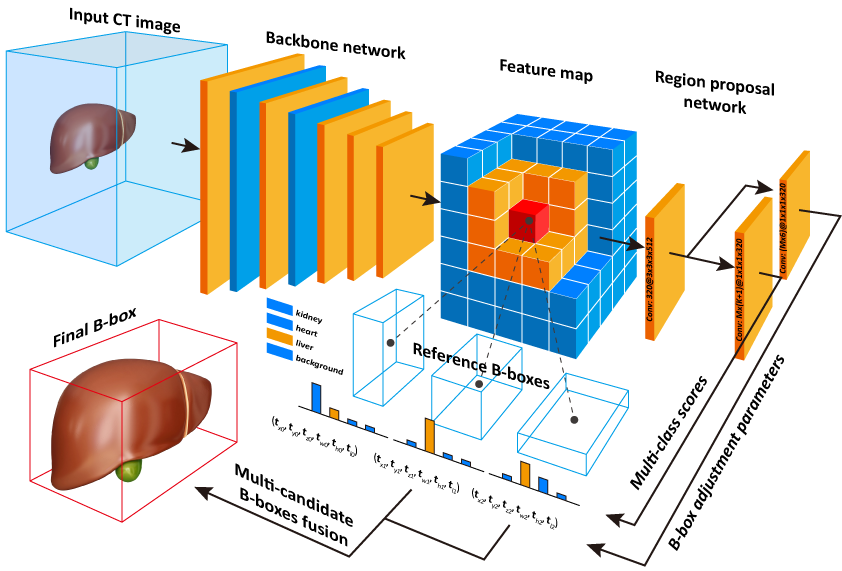
This is a modified version of Caffe which supports the 3D Faster R-CNN framework and 3D Region Proposal Network as described in our paper [Efficient Multiple Organ Localization in CT Image using 3D Region Proposal Network](Early access on IEEE Transactions on Medical Imaging).
This code has been compiled and passed on Windows 7 (64 bits) using Visual Studio 2013.
Requirements: Visual Studio 2013, ITK-4.10, CUDA 8.0 and cuDNN v5
Please make sure CUDA and cuDNN have been installed correctly on your computer.
Clone the project by running:
git clone https://github.com/superxuang/caffe_3d_faster_rcnn.git
In .\windows\Caffe.bat set ITK_PATH to ITK intall path (the path containing ITK include,lib folders).
Run .\windows\Caffe.bat and build the project caffe in Visual Studio 2013.
Please download and unzip the CT images from LiTS challenge. Note that, the original CT images of LiTS dataset are stored in *.nii format. Please convert them to *.mhd format.
The organ bounding-box annotations could be downloaded from this repository or IEEE DataPort.
Move the CT images and the bounding-box annotations to a data folder, and create an entry list file (_train_set_list.txt) in the same folder. To this end, the data folder is organized in the folloing way:
└── data folder
├── _train_set_list.txt
├── volume-0.mhd
├── volume-0.raw
├── segmentation-0.mhd
├── segmentation-0.raw
├── segmentation-0.txt
├── volume-1.mhd
├── volume-1.raw
├── segmentation-1.mhd
├── segmentation-1.raw
├── segmentation-1.txt
| .......................
├── volume-130.mhd
├── volume-130.raw
├── segmentation-130.mhd
├── segmentation-130.raw
└── segmentation-130.txt
The entry list file _train_set_list.txt stores the filenames that are actually used for training. Each line of the entry list file corresponds a data sample. Here is an example of the entry list file _train_set_list.txt corresponding to above data folder. Note that, the segmentation mask file (segmentation-N.mhd and segmentation-N.raw) is not necessary for neither training nor testing. We just reserve it for further research (e.g. organ segmentation). You could just make the segmentation mask files absent and fill the second column of the entry list file with a non-existent filename.
volume-0.mhd segmentation-0.mhd segmentation-0.txt
volume-1.mhd segmentation-1.mhd segmentation-1.txt
.......................
volume-130.mhd segmentation-130.mhd segmentation-130.txt
Modify the path parameter of datalayer in .\models\ours\net.prototxt.
layer {
name: 'mhd_input'
type: 'MHDRoiData'
top: 'data'
top: 'im_info'
top: 'gt_boxes'
mhd_data_param {
source: "F:/Deep/MyDataset/LITS/_train_set_list.txt" # the entry list file mentioned above
root_folder: "F:/Deep/MyDataset/LITS/" # the data folder mentioned above
batch_size: 1
shuffle: true
hist_matching: false
truncate_probability: 0.5
min_truncate_length: 50
inplane_shift: 5
min_intensity: -1000
max_intensity: 1600
random_deform: 0.0
deform_control_point: 2
deform_sigma: 15.0
contour_name_list {
name: "liver"
name: "lung-r"
name: "lung-l"
name: "kidney-r"
name: "kidney-l"
name: "femur-r"
name: "femur-l"
name: "bladder"
name: "heart"
name: "spleen"
name: "pancreas"
}
resample_spacing_x: 2
resample_spacing_y: 2
resample_spacing_z: 2
max_width: 150
max_height: 150
max_length: 1000
}
include: { phase: TRAIN }
}
Run .\models\ours\train.bat
Please cite our paper and Caffe if it is useful for your research:
@article{xu2019organlocalization,
author = {Xu, Xuanang and Zhou, Fugen and Liu, Bo and Fu, Dongshan and Bai, Xiangzhi},
journal = {IEEE Transactions on Medical Imaging},
title = {Efficient Multiple Organ Localization in CT Image using 3D Region Proposal Network},
year = {2019}
doi = {10.1109/TMI.2019.2894854},
volume = {},
number = {},
pages = {1-1},
ISSN = {0278-0062},
month = {},
}
@article{jia2014caffe,
author = {Jia, Yangqing and Shelhamer, Evan and Donahue, Jeff and Karayev, Sergey and Long, Jonathan and Girshick, Ross and Guadarrama, Sergio and Darrell, Trevor},
journal = {arXiv preprint arXiv:1408.5093},
title = {Caffe: Convolutional Architecture for Fast Feature Embedding},
year = {2014}
}
Caffe is released under the BSD 2-Clause license. The BVLC reference models are released for unrestricted use.