In clinical practice, multi-modal magnetic resonance imaging (MRI)
with different contrasts is usually acquired in a single study
to assess different properties of the same region of interest in human body.
The whole acquisition process can be accelerated
by having one or more modalities under-sampled in the
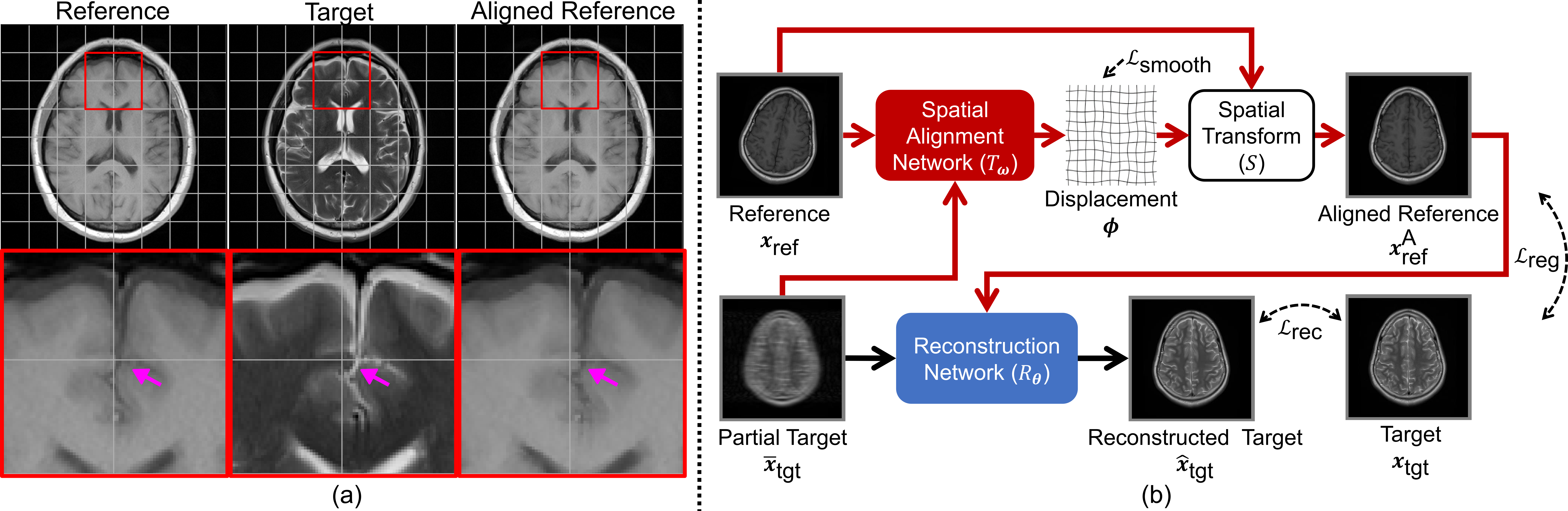 The above figure is a real case demonstrating
the existence of spatial misalignment (a), and the overview of the proposed method (b).
In (a), a real case of multi-modal
MRI acquired for the diagnostic purpose demonstrates the existence
of spatial misalignment (highlighted by arrows) between the reference
(T1-weighted) and the target (T2-weighted) images.
The aligned reference image is also available to show
the effect of our proposed spatial alignment network.
In (b), a spatial alignment network is integrated into the multi-modal MRI reconstruction pipeline
to compensate for the spatial misalignment between the fully-sampled
reference image and the under-sampled target. The data flow for the
conventional deep-learning-based reconstruction is shown in black
arrows; and the red arrows are for additional data flow related to
our proposed spatial alignment network.
The above figure is a real case demonstrating
the existence of spatial misalignment (a), and the overview of the proposed method (b).
In (a), a real case of multi-modal
MRI acquired for the diagnostic purpose demonstrates the existence
of spatial misalignment (highlighted by arrows) between the reference
(T1-weighted) and the target (T2-weighted) images.
The aligned reference image is also available to show
the effect of our proposed spatial alignment network.
In (b), a spatial alignment network is integrated into the multi-modal MRI reconstruction pipeline
to compensate for the spatial misalignment between the fully-sampled
reference image and the under-sampled target. The data flow for the
conventional deep-learning-based reconstruction is shown in black
arrows; and the red arrows are for additional data flow related to
our proposed spatial alignment network.
For more details on the proposed method, please refer to https://ieeexplore.ieee.org/document/9745968.
Store data in h5 files
- Unzip fastMRI brain DICOM to
fastMRI_brain_DICOMfolder. - Convert all dicom to
brain_niifolder.
ls fastMRI_brain_DICOM | while read X; \
do XX="brain_nii/${X}"; mkdir ${XX}; \
echo "dcm2niix -z n -f '%j-%p' -o ${XX} fastMRI_brain_DICOM/${X} 2>${XX}/error.log 1>${XX}/out.log"; \
done | parallel --bar- Convert selected nii to h5.
# T1 weighted
cat t1_t2_paired_6875_{train,val,test}.csv | cut -f1 -d ',' | while read x; \
do python3 convert_fastMRIDICOM.py "${x%.h5}.nii" "${x}" T1; \
done
# T2 weighted
cat t1_t2_paired_6875_{train,val,test}.csv | cut -f2 -d ',' | while read x; \
do python3 convert_fastMRIDICOM.py "${x%.h5}.nii" "${x}" T2;
doneRun commands_train_eval.sh to start training and evaluation of methods.
@ARTICLE{9745968,
author={Xuan, Kai and Xiang, Lei and Huang, Xiaoqian and Zhang, Lichi and Liao, Shu and Shen, Dinggang and Wang, Qian},
journal={IEEE Transactions on Medical Imaging},
title={Multi-Modal MRI Reconstruction Assisted with Spatial Alignment Network},
year={2022},
volume={},
number={},
pages={1-1},
doi={10.1109/TMI.2022.3164050}}