scHolography is an neural network-based computational toolkit for integrative scRNA-seq and ST data analysis. Our pipeline enables 3D spatial inference at a high resolution. Instead of mapping cells to a spot on the fixed ST slice, our reconstruction result is orientation-free, and the inferred structure will be varied for different scRNA-seq data inputs. Together with our downstream analytical functions, we aim to bring new perspectives on scRNA-seq and ST data for researchers.
The deep learning functionalities of our package powers by the Keras API. To install Keras:
if (!requireNamespace("remotes", quietly = TRUE)) {
install.packages("remotes")
}
remotes::install_github(sprintf("rstudio/%s", c("reticulate", "tensorflow", "keras")))
reticulate::miniconda_uninstall()
reticulate::install_miniconda()
tensorflow::install_tensorflow(method = 'conda',envname = 'r-tensorflow',version = '2.12',conda_python_version = '3.9')
######### After above steps, for Mac Apple silicon users, you need to install tensorflow-macos instead of tensorflow by first activating the conda environment and then installing tensorflow-macos in your terminal
## conda activate /Users/YOURPATH/Library/r-miniconda/envs/r-tensorflow
## python -m pip install tensorflow-macos==2.12.0
######### Then you can set the RETICULATE_PYTHON to the python in the conda environment in your R/RStudio
## Sys.setenv(RETICULATE_PYTHON = "/Users/YOURPATH/Library/r-miniconda/envs/r-tensorflow/bin/python")
reticulate::use_condaenv('r-tensorflow')To confirm if the installation is successful:
library(tensorflow)
tf$constant("Hello Tensorflow!")Successful installation will give the output:
tf.Tensor(b'Hello Tensorflow!', shape=(), dtype=string)
DO NOT PROCEED TO THE NEXT STEP IF KERAS INSTALLATION FAILS. For any Keras installation issues, please refer to the Keras page: https://github.com/rstudio/keras
The default integration and SC and ST objects are based on Seurat V4. To install compatible Seurat V4:
options(repos = c("https://satijalab.r-universe.dev", getOption("repos")))
remotes::install_version("SeuratObject", "4.1.3")
remotes::install_version("Seurat", "4.3.0", upgrade = FALSE) remotes::install_github("YiLab-SC/scHolography", upgrade = FALSE)Load scHolography for use:
library(scHolography)For demonstration, we are using our in-house human skin mouse brain data. The pocessed Seurat objects can be downloaded from:
The human skin scRNA-seq data: https://drive.google.com/file/d/1J-TKEcv5Lmom19mSSFLfgnI9JnBtW_Au/view?usp=sharing
The human skin ST 10X Visium data: https://drive.google.com/file/d/1aF0zv5uEHu-42BJQWXtkLy7qrf9ELfq9/view?usp=sharing
Load packages:
library(scHolography)
library(Seurat)
library(ggplot2)
library(dplyr)
library(RColorBrewer)
library(viridis)Load data into our workspace:
low.res.sp<-readRDS("~/Downloads//low.res.human.sk.rds")
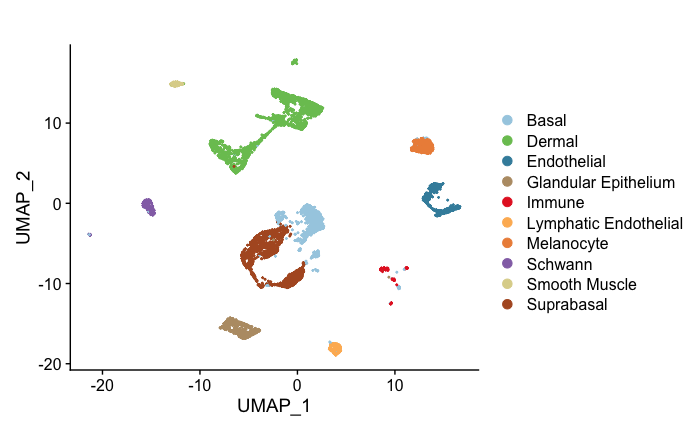
high.res.sp<-readRDS("~/Downloads//high.res.human.sk.rds")Seurat::DimPlot(high.res.sp,group.by = "celltype",cols = c("#A6CEE3","#79C360", "#3F8EAA", "#B89B74", "#E52829", "#FDB762", "#ED8F47", "#9471B4" ,"#DDD399" ,"#B15928"))+ggplot2::ggtitle("")scHolography can take in objects directly after quality control and automatically perform normalization before integration. dataAlign integrations expression data from scRNA-seq and ST modalities. trainHolography trains neural network models and infers a SMN graph.
sp.integrated <- dataAlign(low.res.sp,high.res.sp,nPCtoUse = 32,scProcessed = T)
scHolography.obj<-trainHolography(sp.integrated,n.slot = 30,n.pcUse = 32,n.pcOut = 32,n.repeat = 30)The reconstruction results can be visualized in 3D with scHolographyPlot:
### Set the celltype as factor
scHolography.obj$scHolography.sc$celltype<- factor(as.character(scHolography.obj$scHolography.sc$celltype),levels = c("Basal" ,"Endothelial","Dermal", "Glandular Epithelium","Immune" , "Lymphatic Endothelial" ,"Melanocyte" ,"Schwann" ,"Smooth Muscle" , "Suprabasal" ))
### To orient the plot in a more intuitive way
scHolography.obj$scHolography.sc$z3d_sp <- -scHolography.obj$scHolography.sc$z3d_sp
scHolography.obj$scHolography.sc$x3d_sp <- -scHolography.obj$scHolography.sc$x3d_sp
scHolography.obj$scHolography.sc$y3d_sp <- -scHolography.obj$scHolography.sc$y3d_sp
scene = list(camera = list(eye = list(x = -1, z = 0, y = 2)))
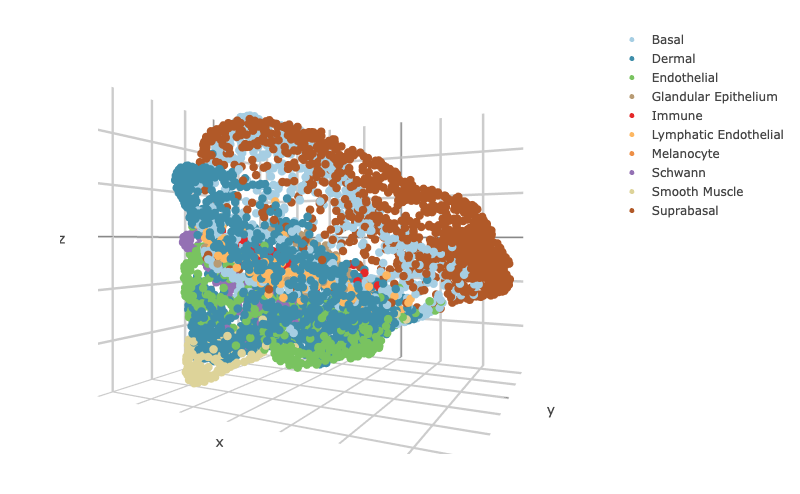
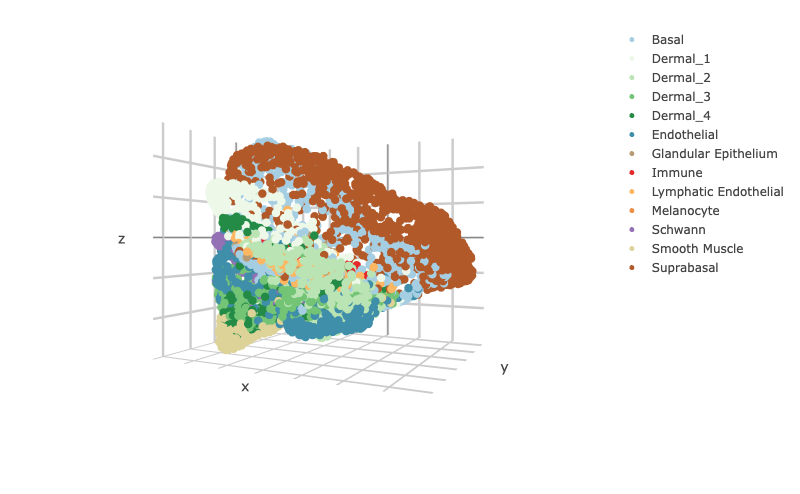
### To visualize the 3D structure colored by celltype
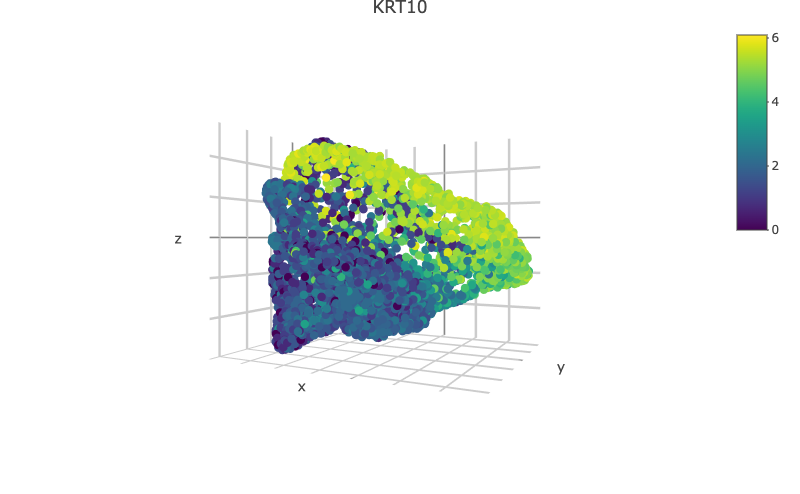
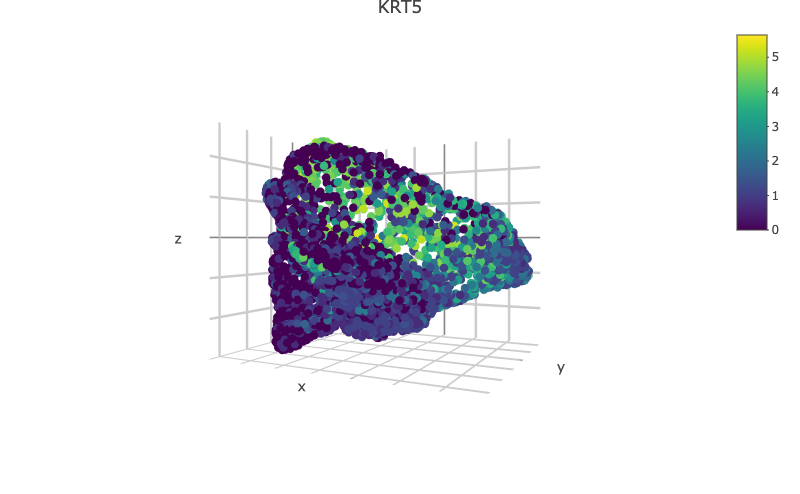
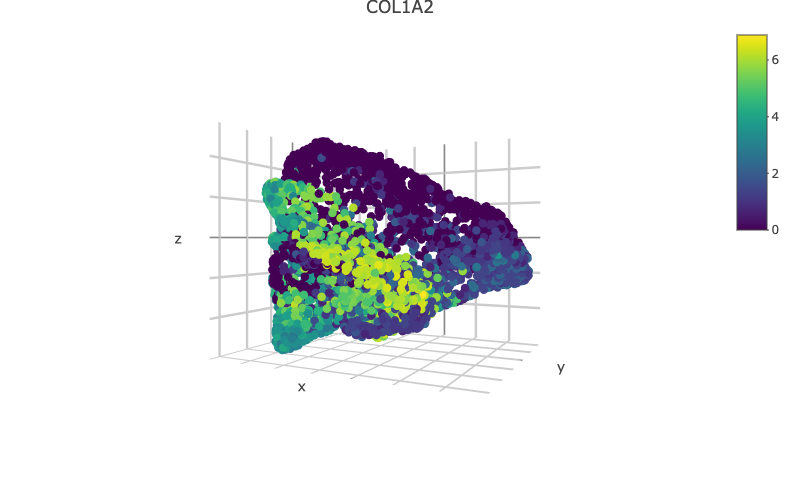
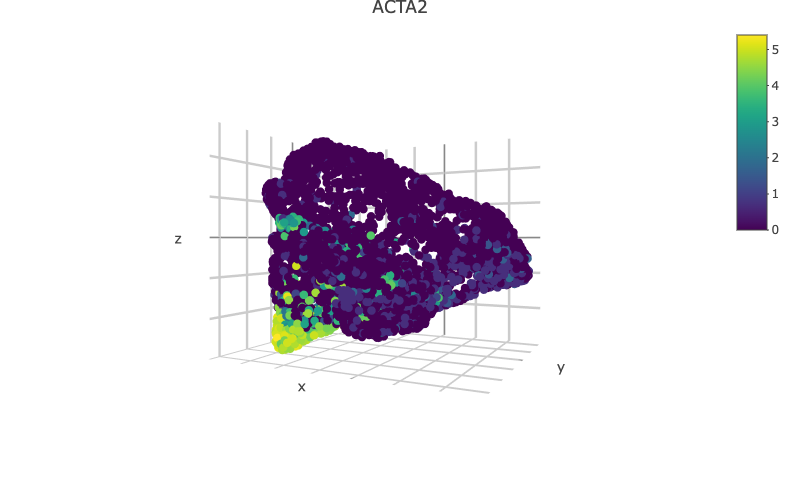
scHolographyPlot(scHolography.obj,color.by = "celltype")%>% plotly::layout(scene = scene)scHolography also assists visualizations of different features on the reconstructed 3D structure. Here, we show three layer neuron marker expression.
scHolographyPlot(scHolography.obj,feature = "KRT10")%>% plotly::layout(scene = scene)scHolographyPlot(scHolography.obj,feature = "KRT5")%>% plotly::layout(scene = scene)scHolographyPlot(scHolography.obj,feature = "COL1A2")%>% plotly::layout(scene = scene)scHolographyPlot(scHolography.obj,feature = "ACTA2")%>% plotly::layout(scene = scene)Based on the reconstructed graph, scHolography can investigate spatial relationship among cell clusters of interest by calculating their SMN distances. Here we demonstrate a boxplot for distances between the major skin cells to all smooth muscle cells.
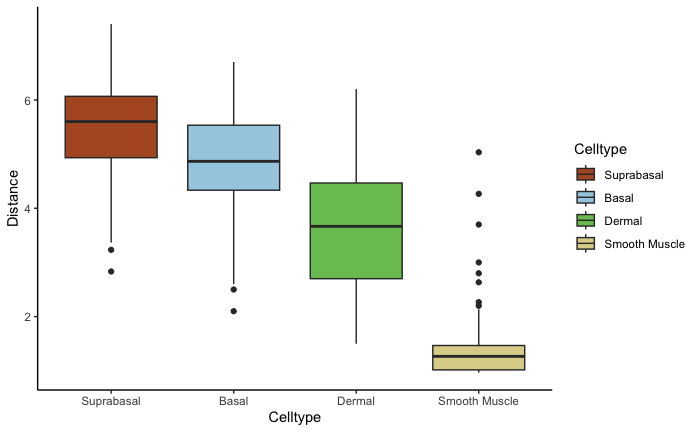
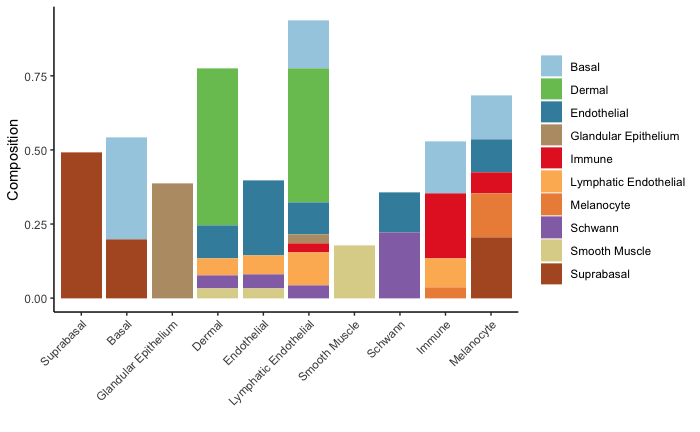
clusterDistanceBoxplot(scHolography.obj,annotationToUse = "celltype",reference.cluster = "Smooth Muscle",query.cluster.list = c("Suprabasal","Basal","Dermal","Smooth Muscle"))For each cell cluster, we can further examine its inferred microenvironment by dissecting their first-degree neighbor cell type composition on the SMN graph. scHolography enables this query with scHolographyNeighborCompPlot. This function output 1. first-degree neighbor composition plot of all neighbors; 2. first-degree neighbor composition plot for only enriched neighboring cell types; 3. significance levels for each enriched neighboring cell type.
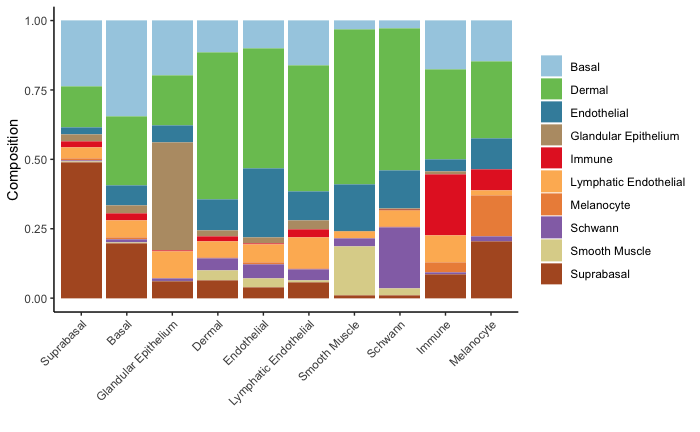
neighbor.comp<- scHolographyNeighborCompPlot(scHolography.obj,annotationToUse = "celltype",query.cluster = c("Suprabasal","Basal","Glandular Epithelium","Dermal","Endothelial","Lymphatic Endothelial","Smooth Muscle", "Schwann","Immune","Melanocyte"))
my.color.order= c("#A6CEE3" ,"#79C360", "#3F8EAA" ,colorRampPalette(brewer.pal(12,"Paired"))(10)[4:10] ) # color order for the plot
neighbor.comp$neighbor.comp.plot+scale_fill_manual(values = my.color.order)neighbor.comp$neighbor.comp.sig.plot+scale_fill_manual(values = my.color.order)neighbor.comp$significance$Suprabasal
Suprabasal
0
$Basal
Basal Suprabasal
0.0000000000 0.0002323026
$`Glandular Epithelium`
Glandular Epithelium
0
$Dermal
Dermal Endothelial Lymphatic Endothelial Schwann
0.000000e+00 1.252084e-13 1.381670e-28 3.444694e-29
Smooth Muscle
3.282852e-40
$Endothelial
Endothelial Lymphatic Endothelial Schwann Smooth Muscle
0.000000e+00 3.718200e-02 1.030445e-05 2.715165e-11
$`Lymphatic Endothelial`
Basal Dermal Endothelial Glandular Epithelium
2.298314e-10 1.070546e-06 1.277586e-11 1.084878e-17
Immune Lymphatic Endothelial Schwann
2.852786e-10 8.049785e-148 8.850146e-05
$`Smooth Muscle`
Smooth Muscle
3.447845e-194
$Schwann
Endothelial Schwann
4.107800e-02 8.314729e-259
$Immune
Basal Immune Lymphatic Endothelial Melanocyte
2.309712e-06 2.654720e-235 8.292197e-06 2.322086e-07
$Melanocyte
Basal Endothelial Immune Melanocyte Suprabasal
4.217856e-04 1.134285e-02 4.239131e-26 1.380532e-155 2.910273e-03
The findSpatialNeighborhood function aims to define distinct spatial neighborhoods and study single-cell spatial heterogeneity in a transcriptome-spatial integrated manner. First, the function decides the number of distinct neighborhoods to define from scHolography inferred query cell spatial distribution. The silhouette coefficient optimizes the number of spatial neighborhoods. The accumulated SMN expression profile of SMNs for each query cell is defined as the sum of the scRNA-seq count of all SMNs of the query cell. The accumulated SMN expression matrix is normalized and the spatial neighborhoods are defined using K-means clustering with the optimized cluster number or by setting the nNeighborhood. Differentially expressed genes are found for both accumulated SMN and single-cell expressions of each spatial neighborhood. In this example, we investigate the spatial neighborhood of human dermal cells.
#Find the spatial neighborhood of Dermal cells
spatial.neighbor.Dermal <- findSpatialNeighborhood(scHolography.obj ,annotationToUse = "celltype",query.cluster = c("Dermal"),orig.assay = "RNA",nNeighborhood = 4)
#Rename the spatial neighborhood and Define the color for the plot
spatial.neighbor.Dermal$scHolography.obj$scHolography.sc$spatial.neighborhood[which(spatial.neighbor.Dermal$scHolography.obj$scHolography.sc$spatial.neighborhood%in%c(as.character(1:4)))] <- paste0("Dermal_",spatial.neighbor.Dermal$scHolography.obj$scHolography.sc$spatial.neighborhood[which(spatial.neighbor.Dermal$scHolography.obj$scHolography.sc$spatial.neighborhood%in%c(as.character(1:4)))])
fib.sp.col <- c(c(colorRampPalette(brewer.pal(12,"Paired"))(10)[1],(brewer.pal(4,"Greens"))),colorRampPalette(brewer.pal(12,"Paired"))(10)[c(2,4:10)])
#Plot the spatial neighborhood
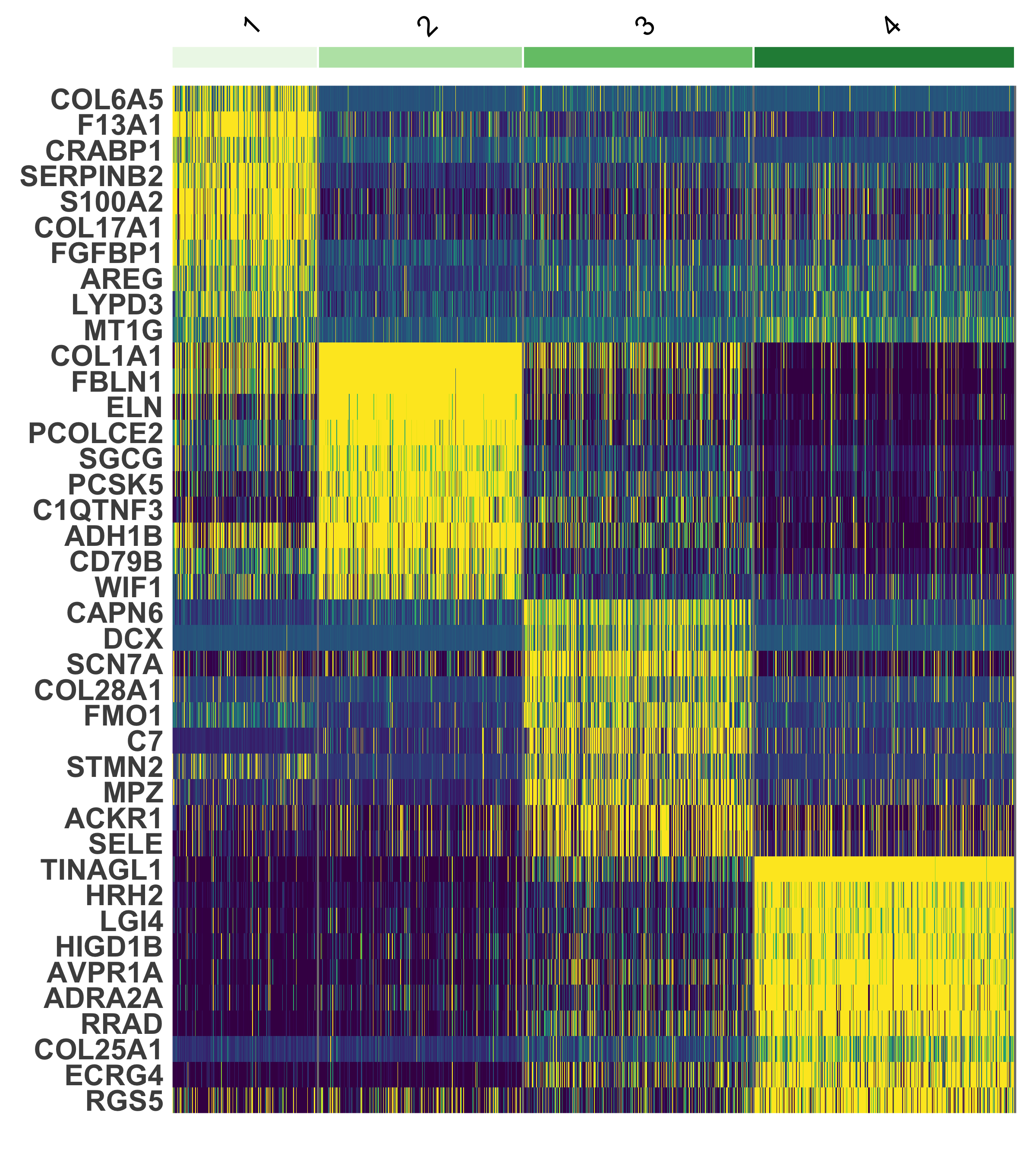
scHolography::scHolographyPlot(spatial.neighbor.Dermal$scHolography.obj,color.by = "spatial.neighborhood",color = fib.sp.col)%>% plotly::layout(scene = scene)We can visualize the expression of the top 10 differentially expressed genes for accumulated SMN in each spatial neighborhood of Dermal cells.
spatial.neighbor.Dermal$neighbor.marker %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log2FC) -> top10
Seurat::DoHeatmap(spatial.neighbor.Dermal$bulk.count.obj,assay = "SCT", features = top10$gene,group.colors = brewer.pal(4,"Greens")) + NoLegend()+scale_fill_viridis()+theme(axis.text = element_text(size = 16,face = "bold"))sessionInfo()R version 4.2.1 (2022-06-23)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 14.4.1
Matrix products: default
LAPACK: /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] RColorBrewer_1.1-3 dplyr_1.1.2 ggplot2_3.4.2 SeuratObject_4.1.3 Seurat_4.3.0
[6] scHolography_0.1.0
loaded via a namespace (and not attached):
[1] Rtsne_0.16 colorspace_2.1-0 deldir_1.0-6 ellipsis_0.3.2
[5] ggridges_0.5.4 base64enc_0.1-3 rstudioapi_0.14 spatstat.data_3.0-1
[9] farver_2.1.1 leiden_0.4.3 listenv_0.9.0 matchingR_1.3.3
[13] ggrepel_0.9.3 fansi_1.0.4 codetools_0.2-19 splines_4.2.1
[17] knitr_1.42 polyclip_1.10-4 zeallot_0.1.0 jsonlite_1.8.4
[21] ica_1.0-3 cluster_2.1.4 png_0.1-8 tfruns_1.5.1
[25] uwot_0.1.14 shiny_1.7.4 sctransform_0.3.5 spatstat.sparse_3.0-1
[29] compiler_4.2.1 httr_1.4.5 Matrix_1.5-4 fastmap_1.1.1
[33] lazyeval_0.2.2 limma_3.52.4 cli_3.6.1 later_1.3.1
[37] htmltools_0.5.5 tools_4.2.1 igraph_1.4.2 gtable_0.3.3
[41] glue_1.6.2 RANN_2.6.1 reshape2_1.4.4 gmodels_2.18.1.1
[45] Rcpp_1.0.10 scattermore_0.8 vctrs_0.6.2 gdata_2.18.0.1
[49] spatstat.explore_3.1-0 nlme_3.1-162 progressr_0.13.0 crosstalk_1.2.0
[53] lmtest_0.9-40 spatstat.random_3.1-4 xfun_0.39 stringr_1.5.0
[57] globals_0.16.2 mime_0.12 miniUI_0.1.1.1 lifecycle_1.0.3
[61] irlba_2.3.5.1 gtools_3.9.4 goftest_1.2-3 future_1.32.0
[65] MASS_7.3-59 zoo_1.8-12 scales_1.2.1 promises_1.2.0.1
[69] spatstat.utils_3.0-2 parallel_4.2.1 yaml_2.3.7 reticulate_1.28-9000
[73] pbapply_1.7-0 gridExtra_2.3 keras_2.11.1 stringi_1.7.12
[77] tensorflow_2.11.0.9000 rlang_1.1.1 pkgconfig_2.0.3 matrixStats_0.63.0
[81] evaluate_0.20 pracma_2.4.2 lattice_0.21-8 ROCR_1.0-11
[85] purrr_1.0.1 tensor_1.5 labeling_0.4.2 patchwork_1.1.2
[89] htmlwidgets_1.6.2 cowplot_1.1.1 tidyselect_1.2.0 parallelly_1.35.0
[93] RcppAnnoy_0.0.20 plyr_1.8.8 magrittr_2.0.3 R6_2.5.1
[97] generics_0.1.3 DBI_1.1.3 withr_2.5.0 pillar_1.9.0
[101] whisker_0.4.1 fitdistrplus_1.1-11 survival_3.5-5 abind_1.4-5
[105] sp_1.6-0 tibble_3.2.1 future.apply_1.10.0 KernSmooth_2.23-20
[109] utf8_1.2.3 spatstat.geom_3.1-0 plotly_4.10.1 rmarkdown_2.21
[113] viridis_0.6.3 grid_4.2.1 data.table_1.14.8 digest_0.6.31
[117] xtable_1.8-4 tidyr_1.3.0 httpuv_1.6.9 munsell_0.5.0
[121] viridisLite_0.4.2
# packages in environment at /Users/yfy6677/Library/r-miniconda-arm64/envs/r-reticulate:
#
# Name Version Build Channel
absl-py 1.4.0 pypi_0 pypi
array-record 0.2.0 pypi_0 pypi
astunparse 1.6.3 pypi_0 pypi
atk-1.0 2.38.0 hcb7b3dd_1 conda-forge
brotlipy 0.7.0 py39h02fc5c5_1005 conda-forge
bzip2 1.0.8 h3422bc3_4 conda-forge
c-ares 1.18.1 h3422bc3_0 conda-forge
ca-certificates 2023.5.7 hf0a4a13_0 conda-forge
cached-property 1.5.2 hd8ed1ab_1 conda-forge
cached_property 1.5.2 pyha770c72_1 conda-forge
cachetools 5.3.0 pypi_0 pypi
cairo 1.16.0 h4741ed9_1015 conda-forge
certifi 2023.5.7 pyhd8ed1ab_0 conda-forge
cffi 1.15.1 py39h7e6b969_3 conda-forge
charset-normalizer 3.1.0 pyhd8ed1ab_0 conda-forge
click 8.1.3 pypi_0 pypi
cryptography 40.0.2 py39he2a39a8_0 conda-forge
dm-tree 0.1.8 pypi_0 pypi
etils 1.2.0 pypi_0 pypi
expat 2.5.0 hb7217d7_1 conda-forge
flatbuffers 23.3.3 pypi_0 pypi
font-ttf-dejavu-sans-mono 2.37 hab24e00_0 conda-forge
font-ttf-inconsolata 3.000 h77eed37_0 conda-forge
font-ttf-source-code-pro 2.038 h77eed37_0 conda-forge
font-ttf-ubuntu 0.83 hab24e00_0 conda-forge
fontconfig 2.14.2 h82840c6_0 conda-forge
fonts-conda-ecosystem 1 0 conda-forge
fonts-conda-forge 1 0 conda-forge
freetype 2.12.1 hd633e50_1 conda-forge
fribidi 1.0.10 h27ca646_0 conda-forge
gast 0.4.0 pypi_0 pypi
gdk-pixbuf 2.42.10 h1ac0d0d_2 conda-forge
gettext 0.21.1 h0186832_0 conda-forge
giflib 5.2.1 h1a8c8d9_3 conda-forge
google-auth 2.17.3 pypi_0 pypi
google-auth-oauthlib 1.0.0 pypi_0 pypi
google-pasta 0.2.0 pypi_0 pypi
googleapis-common-protos 1.59.0 pypi_0 pypi
graphite2 1.3.13 h9f76cd9_1001 conda-forge
graphviz 8.0.5 h10878c0_0 conda-forge
grpcio 1.54.0 pypi_0 pypi
gtk2 2.24.33 h57013de_2 conda-forge
gts 0.7.6 h4b6d4d6_2 conda-forge
h5py 3.6.0 nompi_py39hd982b79_100 conda-forge
harfbuzz 7.2.0 h46e5fef_0 conda-forge
hdf5 1.12.1 nompi_hd9dbc9e_104 conda-forge
icu 72.1 he12128b_0 conda-forge
idna 3.4 pyhd8ed1ab_0 conda-forge
importlib-metadata 6.6.0 pypi_0 pypi
importlib-resources 5.12.0 pypi_0 pypi
jax 0.4.8 pypi_0 pypi
keras 2.12.0 pypi_0 pypi
krb5 1.20.1 h69eda48_0 conda-forge
lcms2 2.15 hd835a16_1 conda-forge
lerc 4.0.0 h9a09cb3_0 conda-forge
libaec 1.0.6 hb7217d7_1 conda-forge
libblas 3.9.0 16_osxarm64_openblas conda-forge
libcblas 3.9.0 16_osxarm64_openblas conda-forge
libclang 16.0.0 pypi_0 pypi
libcurl 8.0.1 heffe338_0 conda-forge
libcxx 16.0.3 h4653b0c_0 conda-forge
libdeflate 1.18 h1a8c8d9_0 conda-forge
libedit 3.1.20191231 hc8eb9b7_2 conda-forge
libev 4.33 h642e427_1 conda-forge
libexpat 2.5.0 hb7217d7_1 conda-forge
libffi 3.4.2 h3422bc3_5 conda-forge
libgd 2.3.3 h939342b_6 conda-forge
libgfortran 5.0.0 12_2_0_hd922786_31 conda-forge
libgfortran5 12.2.0 h0eea778_31 conda-forge
libglib 2.76.2 h24e9cb9_0 conda-forge
libiconv 1.17 he4db4b2_0 conda-forge
libjpeg-turbo 2.1.5.1 h1a8c8d9_0 conda-forge
liblapack 3.9.0 16_osxarm64_openblas conda-forge
libnghttp2 1.52.0 hae82a92_0 conda-forge
libopenblas 0.3.21 openmp_hc731615_3 conda-forge
libpng 1.6.39 h76d750c_0 conda-forge
libprotobuf 3.19.6 hb5ab8b9_0 conda-forge
librsvg 2.56.0 h6c0e662_0 conda-forge
libsqlite 3.41.2 hb31c410_1 conda-forge
libssh2 1.10.0 h7a5bd25_3 conda-forge
libtiff 4.5.0 h4f7d55c_6 conda-forge
libtool 2.4.7 hb7217d7_0 conda-forge
libwebp 1.3.0 h66d6964_0 conda-forge
libwebp-base 1.3.0 h1a8c8d9_0 conda-forge
libxcb 1.13 h9b22ae9_1004 conda-forge
libxml2 2.10.4 h2aff0a6_0 conda-forge
libzlib 1.2.13 h03a7124_4 conda-forge
llvm-openmp 16.0.3 h1c12783_0 conda-forge
markdown 3.4.3 pypi_0 pypi
markupsafe 2.1.2 pypi_0 pypi
ml-dtypes 0.1.0 pypi_0 pypi
ncurses 6.3 h07bb92c_1 conda-forge
numpy 1.23.2 py39h3668e8b_0 conda-forge
oauthlib 3.2.2 pypi_0 pypi
openjpeg 2.5.0 hbc2ba62_2 conda-forge
openssl 3.1.0 h53f4e23_3 conda-forge
opt-einsum 3.3.0 pypi_0 pypi
packaging 23.1 pyhd8ed1ab_0 conda-forge
pandas 2.0.1 py39h6b13a34_1 conda-forge
pango 1.50.14 h9f7e0c6_1 conda-forge
pcre2 10.40 hb34f9b4_0 conda-forge
pillow 9.5.0 py39hfc21214_0 conda-forge
pip 23.1.2 pyhd8ed1ab_0 conda-forge
pixman 0.40.0 h27ca646_0 conda-forge
platformdirs 3.5.0 pyhd8ed1ab_0 conda-forge
pooch 1.7.0 pyha770c72_3 conda-forge
promise 2.3 pypi_0 pypi
protobuf 4.23.0 pypi_0 pypi
psutil 5.9.5 pypi_0 pypi
pthread-stubs 0.4 h27ca646_1001 conda-forge
pyasn1 0.5.0 pypi_0 pypi
pyasn1-modules 0.3.0 pypi_0 pypi
pycparser 2.21 pyhd8ed1ab_0 conda-forge
pydot 1.4.2 py39h2804cbe_3 conda-forge
pyopenssl 23.1.1 pyhd8ed1ab_0 conda-forge
pyparsing 3.0.9 pyhd8ed1ab_0 conda-forge
pysocks 1.7.1 pyha2e5f31_6 conda-forge
python 3.9.16 hea58f1e_0_cpython conda-forge
python-dateutil 2.8.2 pyhd8ed1ab_0 conda-forge
python-tzdata 2023.3 pyhd8ed1ab_0 conda-forge
python_abi 3.9 3_cp39 conda-forge
pytz 2023.3 pyhd8ed1ab_0 conda-forge
readline 8.2 h92ec313_1 conda-forge
requests 2.29.0 pyhd8ed1ab_0 conda-forge
requests-oauthlib 1.3.1 pypi_0 pypi
rsa 4.9 pypi_0 pypi
scipy 1.10.1 py39hba9bd2d_1 conda-forge
setuptools 67.7.2 pyhd8ed1ab_0 conda-forge
six 1.16.0 pyh6c4a22f_0 conda-forge
tensorboard 2.12.3 pypi_0 pypi
tensorboard-data-server 0.7.0 pypi_0 pypi
tensorflow-datasets 4.9.2 pypi_0 pypi
tensorflow-deps 2.10.0 0 apple
tensorflow-estimator 2.12.0 pypi_0 pypi
tensorflow-hub 0.13.0 pypi_0 pypi
tensorflow-macos 2.12.0 pypi_0 pypi
tensorflow-metadata 1.13.1 pypi_0 pypi
termcolor 2.3.0 pypi_0 pypi
tk 8.6.12 he1e0b03_0 conda-forge
toml 0.10.2 pypi_0 pypi
tqdm 4.65.0 pypi_0 pypi
typing-extensions 4.5.0 hd8ed1ab_0 conda-forge
typing_extensions 4.5.0 pyha770c72_0 conda-forge
tzdata 2023c h71feb2d_0 conda-forge
urllib3 1.26.15 pyhd8ed1ab_0 conda-forge
werkzeug 2.3.4 pypi_0 pypi
wheel 0.40.0 pyhd8ed1ab_0 conda-forge
wrapt 1.14.1 pypi_0 pypi
xorg-libxau 1.0.9 h27ca646_0 conda-forge
xorg-libxdmcp 1.1.3 h27ca646_0 conda-forge
xz 5.2.6 h57fd34a_0 conda-forge
zipp 3.15.0 pypi_0 pypi
zlib 1.2.13 h03a7124_4 conda-forge
zstd 1.5.2 hf913c23_6 conda-forge