Sequencing101
Tres bon argumentairer pour l'usage de la genomique en matiere de plante. C'est un article de KeyGene dans le blog nanopore
Genome sequencing has become an indispensable tool in plant breeding research, through which genetic variation of individual varieties at nucleotide precision can be revealed. Plant breeding companies use reference genomes to:
- effectively develop trait-linked markers and discover genes
- perform comparative genomics to elucidate breeding history
- dissect the genetic architecture of important traits and link them to haplotypes
- expand trait discovery approaches beyond SNP-based analysis
The insights obtained using a reference genome enable better and faster selection of important breeding trait values in the breeding process and therefore allow to bring new varieties to the market faster.
Ils ont mis aussi les 4C avec le 4eme C comme etant le cout in contrast avec le computing de PacBio. Tres fun!
Correctness, Completeness, Contiguity and Cost, (also dubbed as the 4C’s) are of main importance with regard to generating reference genomes. High contiguity (i.e. Telomere-to-Telomere) and completeness can already be obtained by isolating High Molecular Weight (HMW) genomic DNA, in combination with (ultra) long Oxford Nanopore sequencing.
Ce qui m'a le plus impressionner c'est le processus de assembly and annotation en une semaine
very impressive
Price Nanopore
https://github.com/sirselim/jetson_nanopore_sequencing
Oxford Nanopore Technology: revolutionizing genomics research in plants
Application
Unleashing the Power of HiFi Sequencing - How the Sequel IIe System Empowers Life Scientists
Computing env
-
SENCKENBERG BIODIVERSITY GENOMICS SYMPOSIUM 21 April 2021
-
Friday, February 12, 2021 | LeafGo: A Workflow That Enables High-Quality Seven-Day Plant Genome Assembly
-
Wednesday, March 25, 2020 | The Evolution of DNA Sequencing Tools
-
Tuesday, July 7, 2020 | From DNA to Discovery – The Steps of SMRT Sequencing
-
Thursday, May 21, 2020 | Understanding Accuracy in DNA Sequencing
-
The Most Wonderful Webinars of the Year
-
Thursday, December 10, 2020 | Sequencing 101: Ploidy, Haplotypes, and Phasing – How to Get More from Your Sequencing Data
-
Wednesday, November 25, 2020 | Better Apple Pies: HiFi Reads are a Perfect Recipe for High-Quality Apple Genome and Pangenome Assemblies
-
Thursday, October 1, 2020 | Webinar: Crops and Corvids get the Pangenome Treatment with HiFi Sequencing
-
Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm
-
PacBio webinar February 6 2021: Lessons from Assembling High-Quality Reference Genomes for Domesticated & Endangered Ruminants | Video
Contigs assembly alone is not enough in this case and we still need scaffolding of the contigs. We use Hi-C for long-range scaffolding and the Bionano optical map for short range scafolding. I will not go in the details here but basically Optical map is crucial for resolving some of the wrong chimeric contigs and it is also very good in ordering and orientating the scaffolds. Hi-C is very good to join contigs to give chromosome size scaffold
Video at this moment checking the final assembly by comparing gap and repeat content using ggplot2 graph
Can use this graph to compare sesamum indicum assemblies from china pangenome for the PhD.
The comparision of chromosome to a high quality reference genome is another way to check and compare genome assemblies. May use this plot to compare Goenbaek and Zhonzhi13 chromosomal scale assemblies in the phD document also
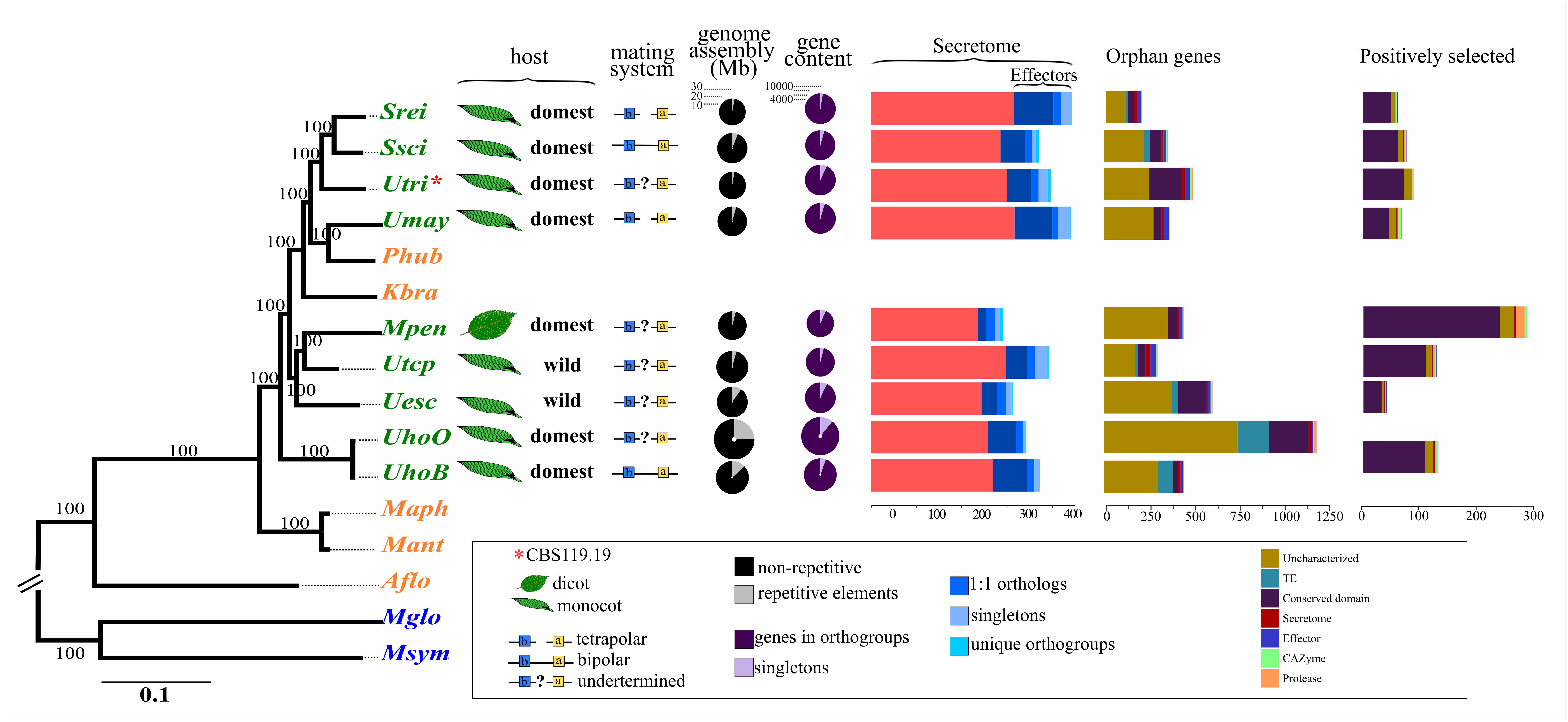
Just randomly get such an inspiring graph from here showing comparative of pathogen genomes. Can be applied to my fungal genome project case.
-
PacBio webinar February 6 2021: Comparative Genomic Analysis in Sorghum Highlights the Extent of Structural Variations | Video
-
Elucidating origins of grapevine agronomic traits with PacBio genome assembly and Iso-Seq analysis
Hifi_assemblers
Hifi free datasets
PacBio Day | Workshop: Whole Genome Sequencing for De Novo Assembly of Any Organism