Official implementation for "Binding-Adaptive Diffusion Models for Structure-Based Drug Design" (AAAI 2024).
Affiliation: Tsinghua University, Peking University, University of Science and Technology of China, University of Chinese Academy of Sciences, ByteDance, Peng Cheng Laboratory.
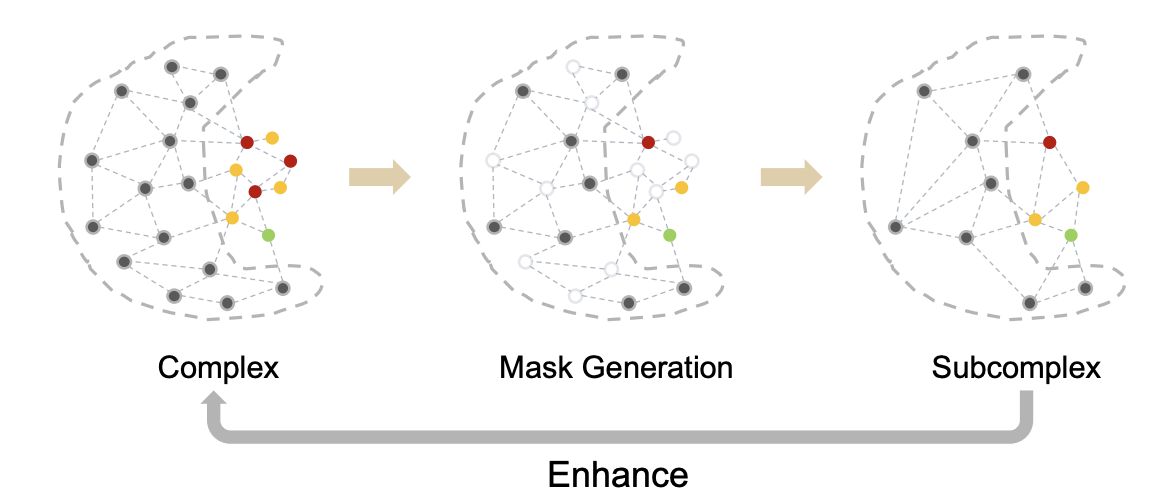 Overview: BindDM extracts subcomplex from protein-ligand complex, and utilizes it to enhance the binding-adaptive 3D molecule generation in complex.
Overview: BindDM extracts subcomplex from protein-ligand complex, and utilizes it to enhance the binding-adaptive 3D molecule generation in complex.
conda env create -f binddm.yaml
conda activate binddmThe data preparation follows TargetDiff. For more details, please refer to the repository of TargetDiff.
python train.pypython sample.pypython evaluate.py@inproceedings{huang2024binddm,
title={Binding-Adaptive Diffusion Models for Structure-Based Drug Design},
author={Huang, Zhilin and Yang, Ling and Zhang, Zaixi and Zhou, Xiangxin and Bao, Yu and Zheng, Xiawu and Yang, Yuwei and Wang, Yu and Yang, Wenming},
booktitle={The AAAI Conference on Artificial Intelligence},
year={2024}
}