LinTIMaT is a statistical method for reconstructing lineages from joint CRISPR-Cas9 mutations and single-cell transcriptomic data. The method is described in the paper titled "Single-cell Lineage Tracing by Integrating CRISPR-Cas9 Mutations with Transcriptomic Data".
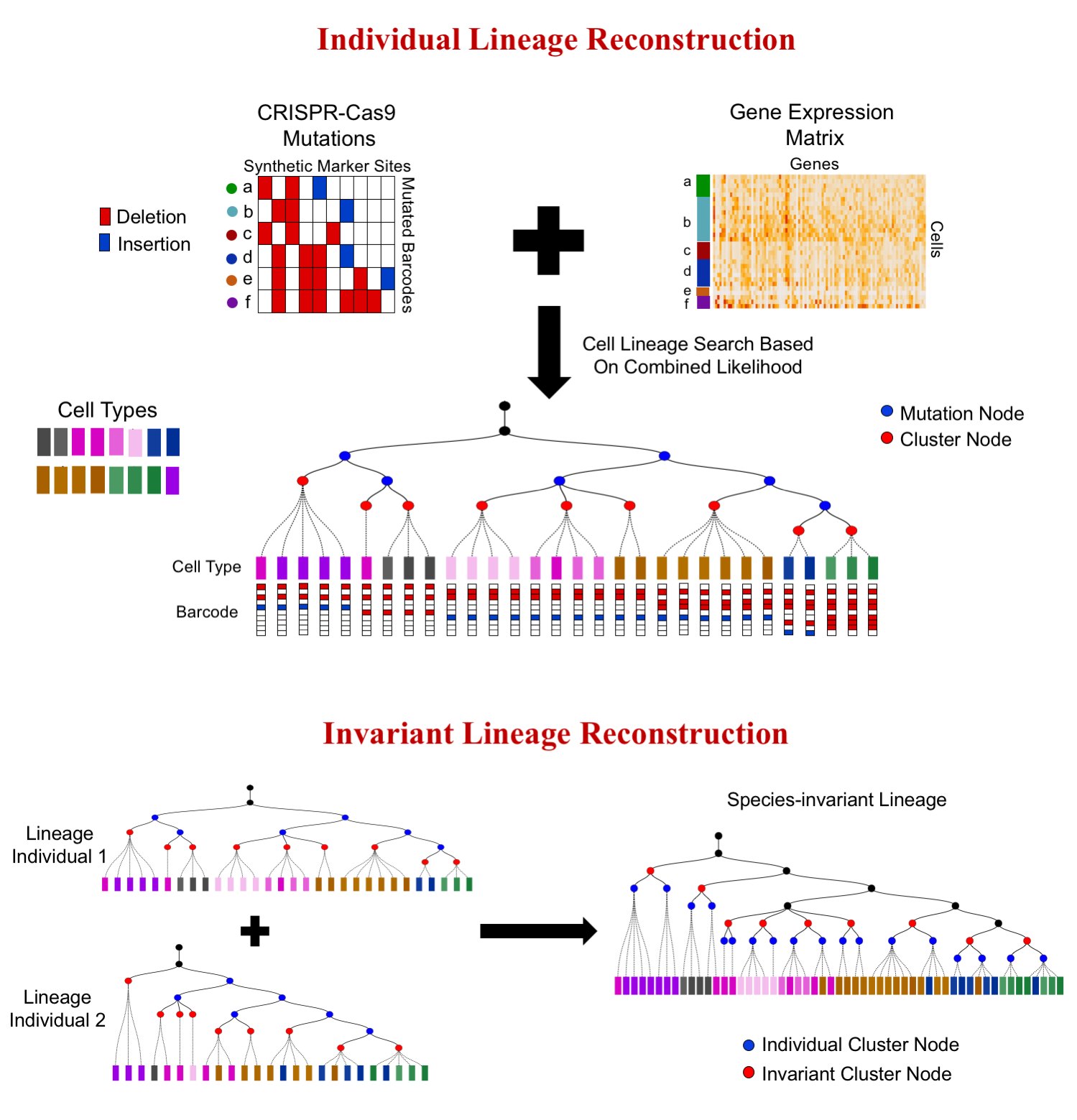
Recent studies combine two novel technologies, single-cell RNA-sequencing and CRISPR-Cas9 barcode editing for elucidating developmental lineages at the whole organism level. To utilize these two complementary data types for lineage tracing, we developed a novel computational method, LinTIMaT. LinTIMaT reconstructs cell lineages in a maximum-likelihood framework by integrating mutation and expression data. Using LinTIMaT, we can reconstruct lineage for a single individual as well as a consensus lineage by combining data from multiple individuals of the same species (see figure below).
See the wiki for details. LinTIMaT can be executed from command line on Mac OS, Windows 10 and Linux machines.