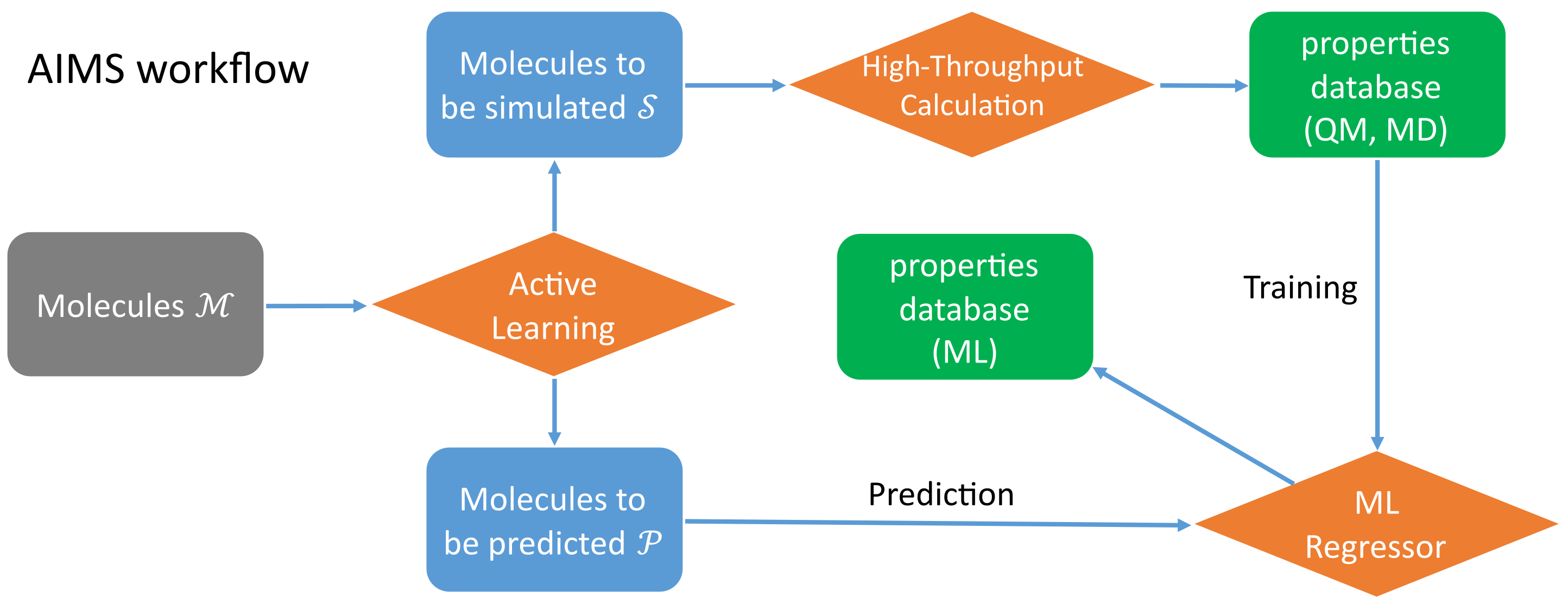
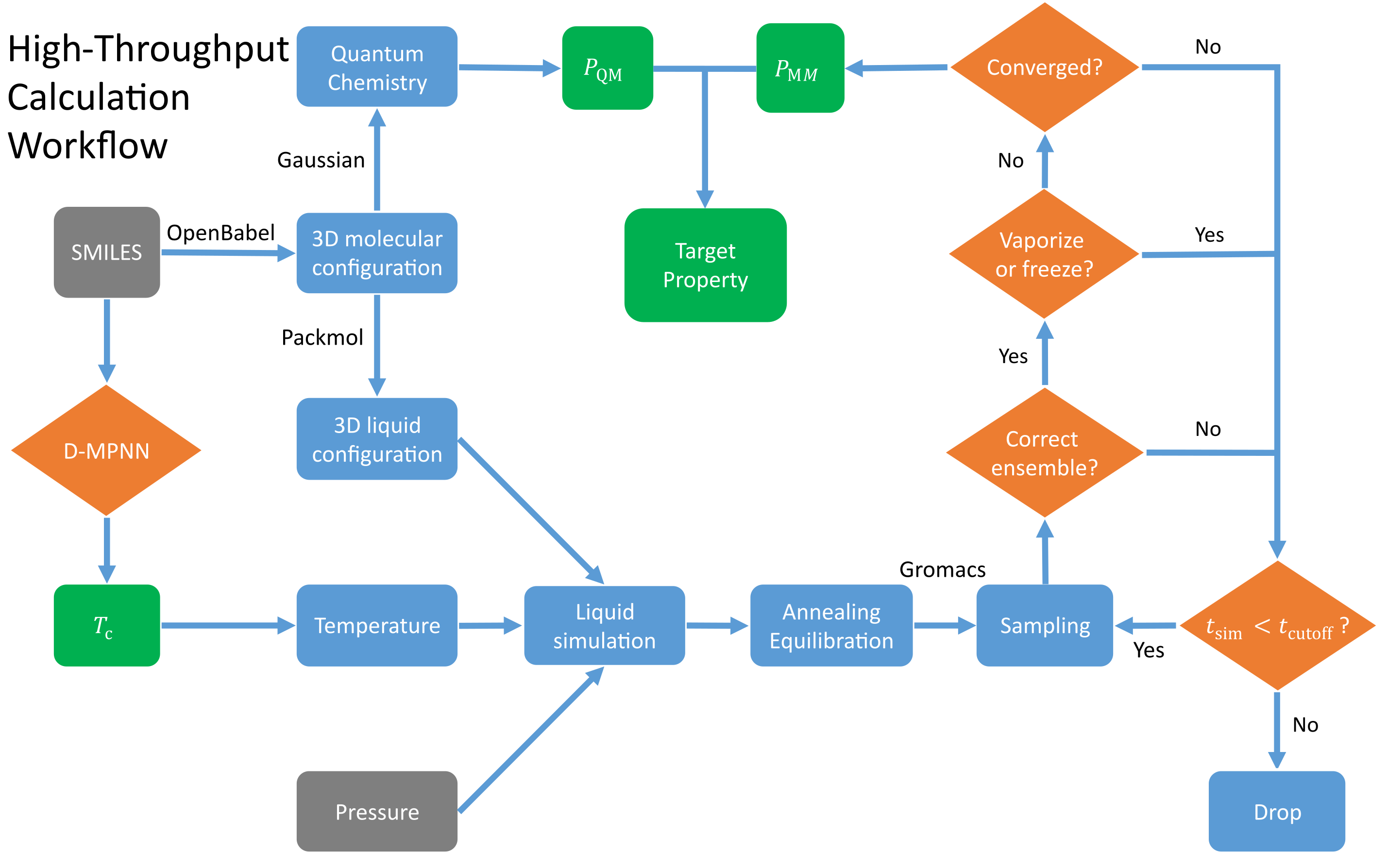
ALMS (Active Learning Molecular Simulation)
ALMS is a program designed to generate large-scale molecular property data sets using machine learning and computational chemistry techniques. Efficient Exploration of Chemical Compound Space Using Active Learning for Prediction of Thermodynamic Properties of Alkane Molecules.
It contains three main parts.
- Active Learning (Gaussian Process Regression, Marginalized Graph Kernel).
- High-throughput Quamtum Chemistry Calculation (GAUSSIAN) and classical Force-Field Molecular Dynamics Simulation (GROMACS).
- Machine Learning Prediction (Direct Message Passing Neural Network).
The workflow of ALMS:
The workflow of high-throughput calculation engine:
Dependencies and Installation
- GAUSSIAN (Quantum Chemistry).
- DFF, Packmol, GROMACS (Molecular Dynamics).
- Require GCC (7.*), NVIDIA Driver and CUDA toolkit(>=10.1) (GraphDot).
conda env create -f environment.yml
conda activate alms
- "monitor.py" need to run on a cluster with SLURM job manager to automatically submit jobs and collect results.
Usages
- Submit molecules to the database.
- Type SMILES of molecules.
python3 submit.py --smiles CCCC CCCCC CCCCCC --files data/test.csv --features_generator rdkit_2d_normalized --heavy_atoms 4 19 - Calculate the kernel matrix and save in data/kernel.pkl. Must run in GPU.
python3 calc_kernel.py --n_jobs 6 - High-throughput QM calculation.
python3 monitor.py --task qm_cv --partition cpu --n_cores 8 --n_jobs 8 --gaussian_exe $GAUSSIAN --n_conformer 1 --stop_uncertainty 0.3 --graph_kernel_type preCalc - High-throughput MD simulation.
python3 monitor.py --task md_npt --partition gtx --n_cores 16 --n_hypercores 32 --n_gpu 2 --n_jobs 8 --packmol_exe $PACKMOL --dff_root $DFF --gmx_exe_analysis gmx_serial --gmx_exe_mdrun gmx_gpu --stop_uncertainty 0.3 --graph_kernel_type preCalc - Export simulation data.
python3 export.py --property cp - Train and predict using D-MPNN.
It is suggested to learn how to use D-MPNN at https://github.com/chemprop/chemprop.
Add temperature as model input by adding
--features_columns T.python3 train.py --data_path cp.csv --dataset_type regression --save_dir ml-models/cp-dmpnn --split_type random --split_sizes 1.0 0.0 0.0 --metric mae --num_fold 1 --ensemble_size 1 --epochs 100 --save_preds --smiles_columns smiles --features_columns T --target_columns cp --features_generator rdkit_2d --num_workers 6 --aggregation sum python3 predict.py --data_path cp_test_set.csv --preds_path cp_preds.csv --checkpoint_dir ml-models/cp-dmpnn --features_generator rdkit_2d --features_columns T