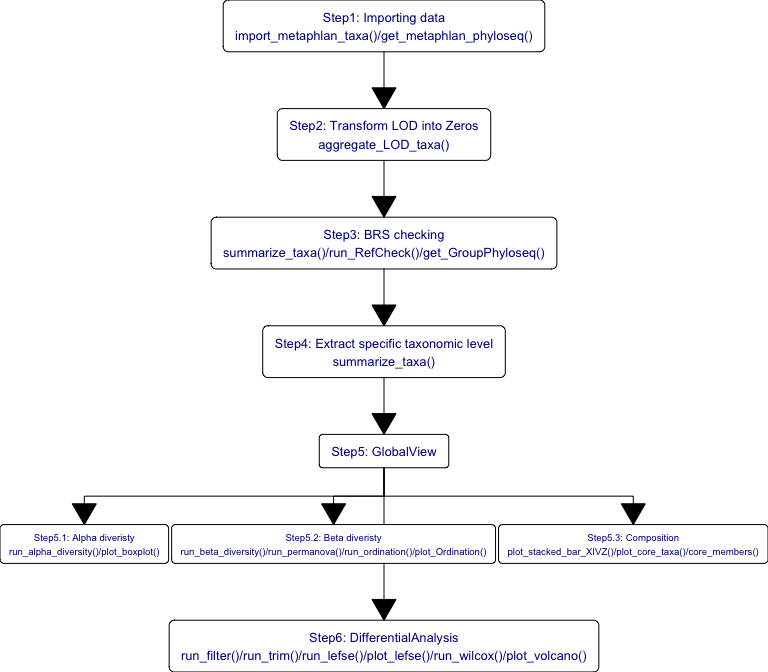
The standard data analysis of 16S and MGS data by XMAS 2.0. The upstream process is performed by in-house pipeline. and this example just shows how to perform downstream data analysis. In briefly, the example comprises the following steps:
-
Converting inputs into phyloseq object;
-
Quality Evaluation;
-
Pre-Processing Data;
-
Diversity analysis;
-
Ordination analysis;
-
Composition analysis;
-
Differential analysis;
-
Network analysis.
Xbiome company
- Submitted to gitlab. (2022-05-28)
- Uploaded bookdwon. (2022-08-04)
- Added Chapter Network Analysis . (2022-08-04)
- Updated bookdwon by XMAS version 2.2.0. (2023-10-27)