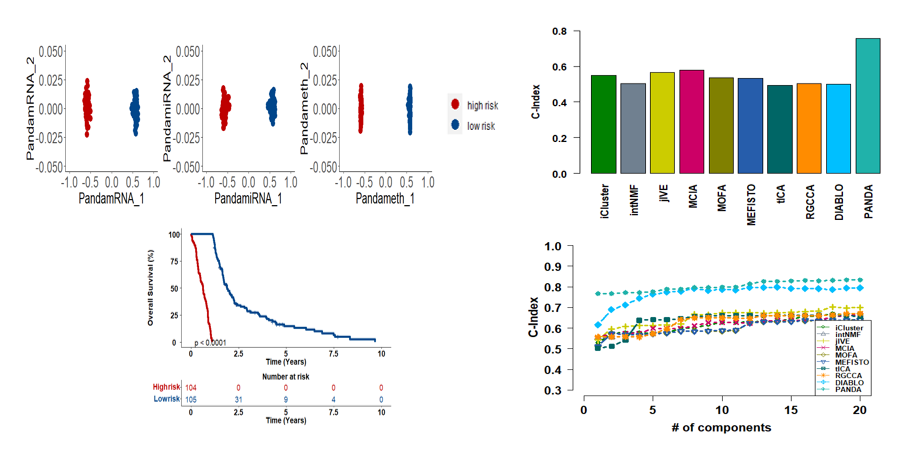
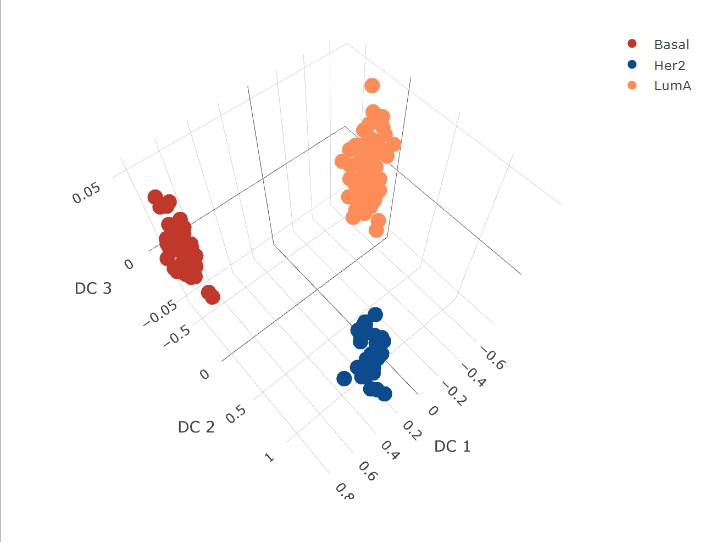
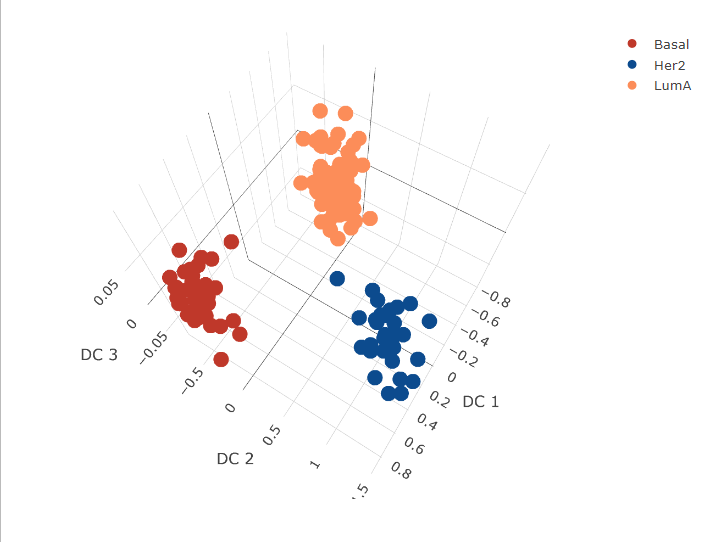
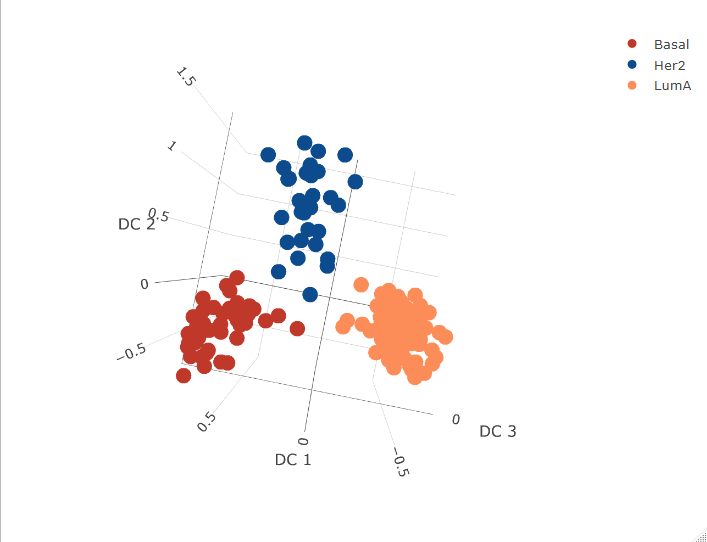
PanDA is a joint discriminant analysis method aimed at fusing multi-omics datasets through finding a discriminant common latent space. PanDA captures cross-omics interaction and consistency and uses an uncorrelated constraint to ensure that the extracted latent components for each omics (omics-specific components) are not highly correlated. Because the components extracted using PanDA contain valuable discriminant information, we refer to them as discriminant components. These components can be used as inputs to several multi-omics analysis tools to enable efficient, improved downstream analysis. Here and in our paper, we demonstrated the advantages of PanDA over ten integrative multi-omics methods through four distinct downstream analyses: single-cell multi-omics data visualization, patient (or tumor) classification, biomarker identification, and clinical outcome prediction.
PanDA can be installed by simply running the following code:
## Install development version
remotes::install_github("WuLabMDA/PANDA")
or by running:
remotes::install_github("muhammadaminu47/PANDA")The input dataset for all the experiments reported in our paper can be found in the ./data/ folder of this repository.
- Basic PanDA example on simulated dataset
- PanDA identifies important markers related to breast cancer
- PanDA application to clinical outcome prediction
The benchmarking results on the simulated, cancer cell line and the TCGA multi-omics datasets can be obtained by running the corresponding code in the ./code/ folder.
An example of the PanDA work flow to get started. Here we used the processed TCGA multiomics dataset from the mixOmics package
library(PANDA)
library(mixOmics) # import the mixOmics library
data(breast.TCGA) # extract the TCGA data
data = list()
data$mirna <- t(breast.TCGA$data.train$mirna)
data$mrna <- t(breast.TCGA$data.train$mrna)
data$protein <- t(breast.TCGA$data.train$protein)
Y <- breast.TCGA$data.train$subtype # use the subtype as the outcome variable
subtype <- factor(Y)
Extract discriminant latent components using PanDA.
labels <- as.numeric(Y)
numComponents <- 10
PanDAModel <- PanDA(data,labels,numComponents)
Plot the discriminant latent representations for the different omics data
mirnaComp <- as.data.frame(DOmicsModel[["DOmicsComponents"]][["mirnaComponents"]])
mrnaComp <- as.data.frame(DOmicsModel[["DOmicsComponents"]][["mrnaComponents"]])
proteinComp <- as.data.frame(DOmicsModel[["DOmicsComponents"]][["proteinComponents"]])
library(plotly)
cols = c('#BF382A', '#0C4B8E', "#fc8d59")
fig1 <- plot_ly(mrnaComp, x = ~`DC 1`, y = ~`DC 2`, z = ~`DC 3`, color = ~subtype, colors = cols)
fig1 <- fig1 %>% add_markers()
fig1 <- fig1 %>% layout(scene = list(xaxis = list(title = 'DC 1'),
yaxis = list(title = 'DC 2'),
zaxis = list(title = 'DC 3')))
fig1
fig2 <- plot_ly(mirnaComp, x = ~`DC 1`, y = ~`DC 2`, z = ~`DC 3`, color = ~subtype, colors = cols)
fig2 <- fig2 %>% add_markers()
fig2 <- fig2 %>% layout(scene = list(xaxis = list(title = 'DC 1'),
yaxis = list(title = 'DC 2'),
zaxis = list(title = 'DC 3')))
fig2
fig3 <- plot_ly(proteinComp, x = ~`DC 1`, y = ~`DC 2`, z = ~`DC 3`, color = ~subtype, colors = cols)
fig3 <- fig3 %>% add_markers()
fig3 <- fig3 %>% layout(scene = list(xaxis = list(title = 'DC 1'),
yaxis = list(title = 'DC 2'),
zaxis = list(title = 'DC 3')))
fig3
For any question, request or bug report please create a new issue in this repository.
We welcome contributions and suggestions from the community. If you have any idea, please submit it as an issue, which we will look into and ask for further explannation if necessary.
PanDA is under continuous development. If you encounter an issue, please make sure you install all the required packages necessary to run the codes. If that does not solve your problem, please open a new issue detailing your encountered problem by providing a code and a demo example. We will try to look into your issue and hopefully provide you with a solution. Thanks.
To cite this work and access a comprehensive description of the PanDA method, please refer to: