Use the live app now. No downloads. No installation. 👇
The GitHub repository is organized into two directories :
-
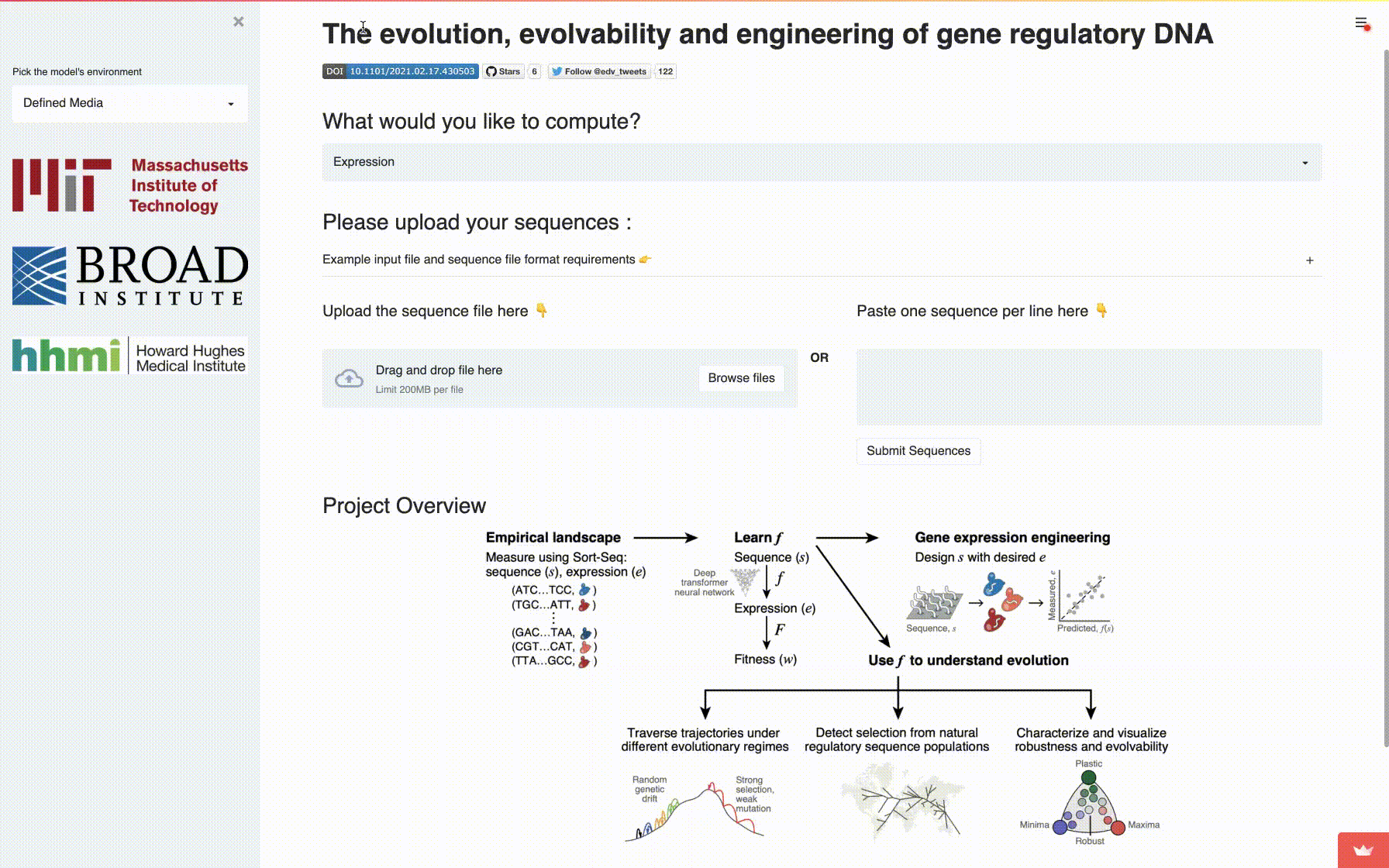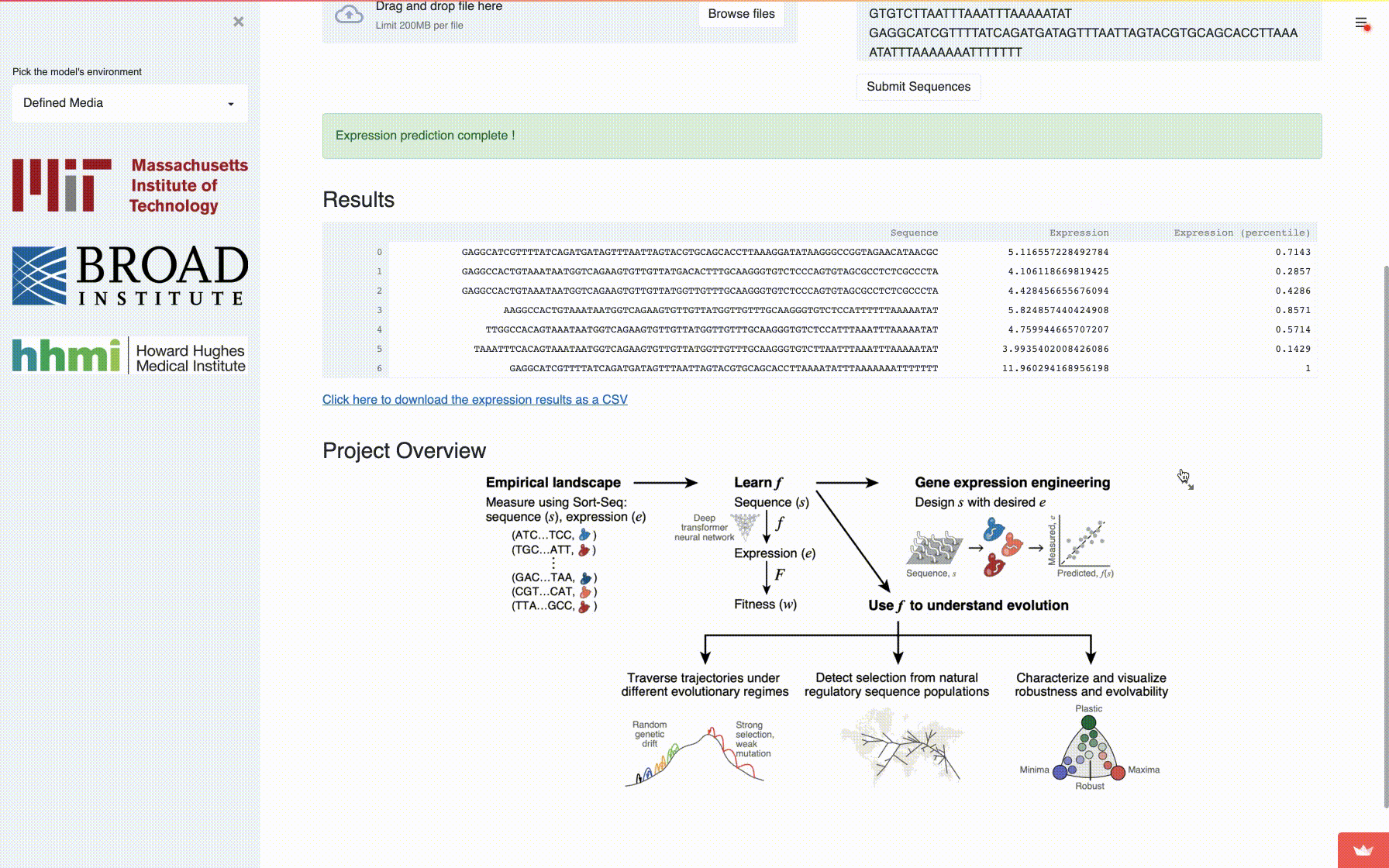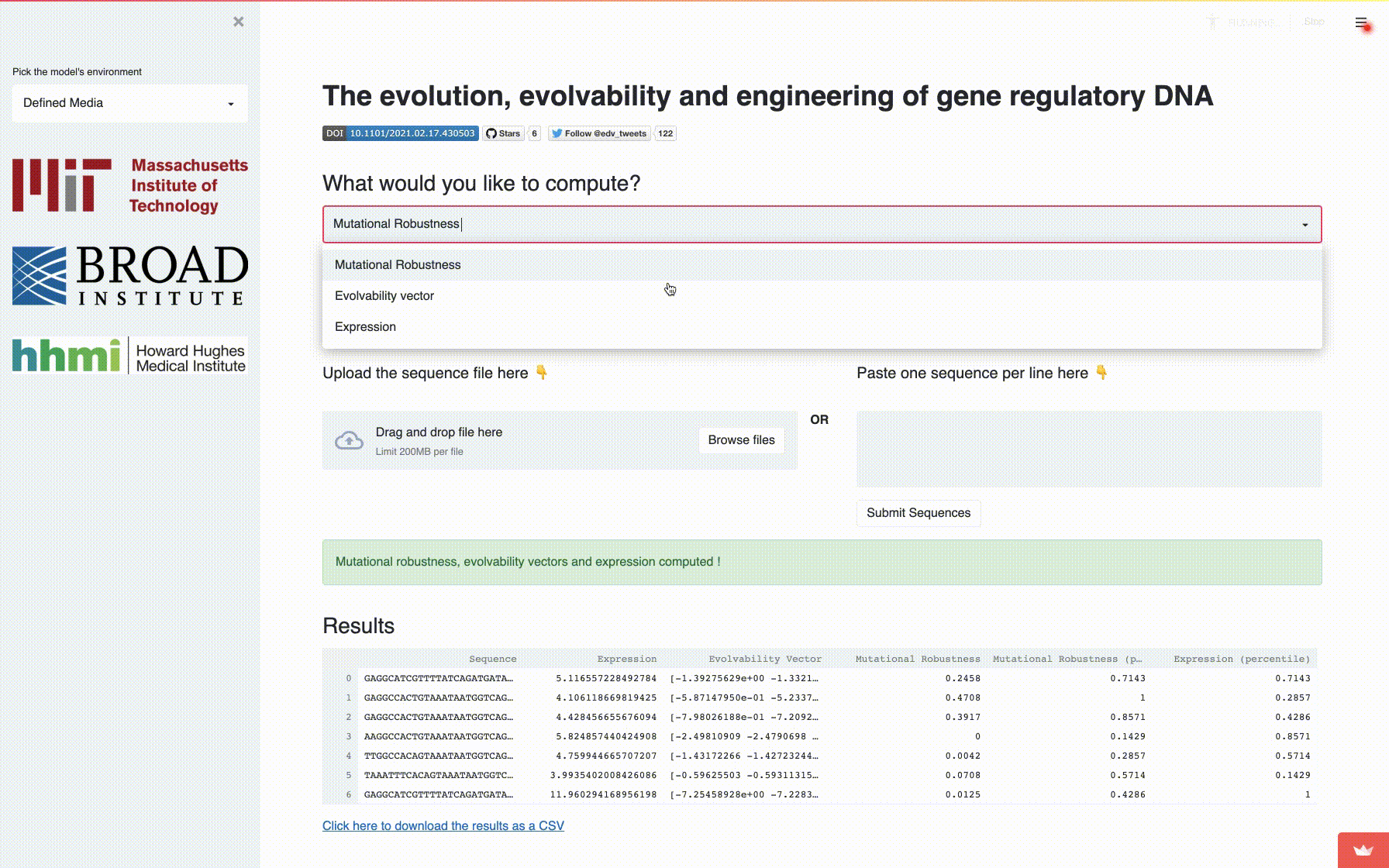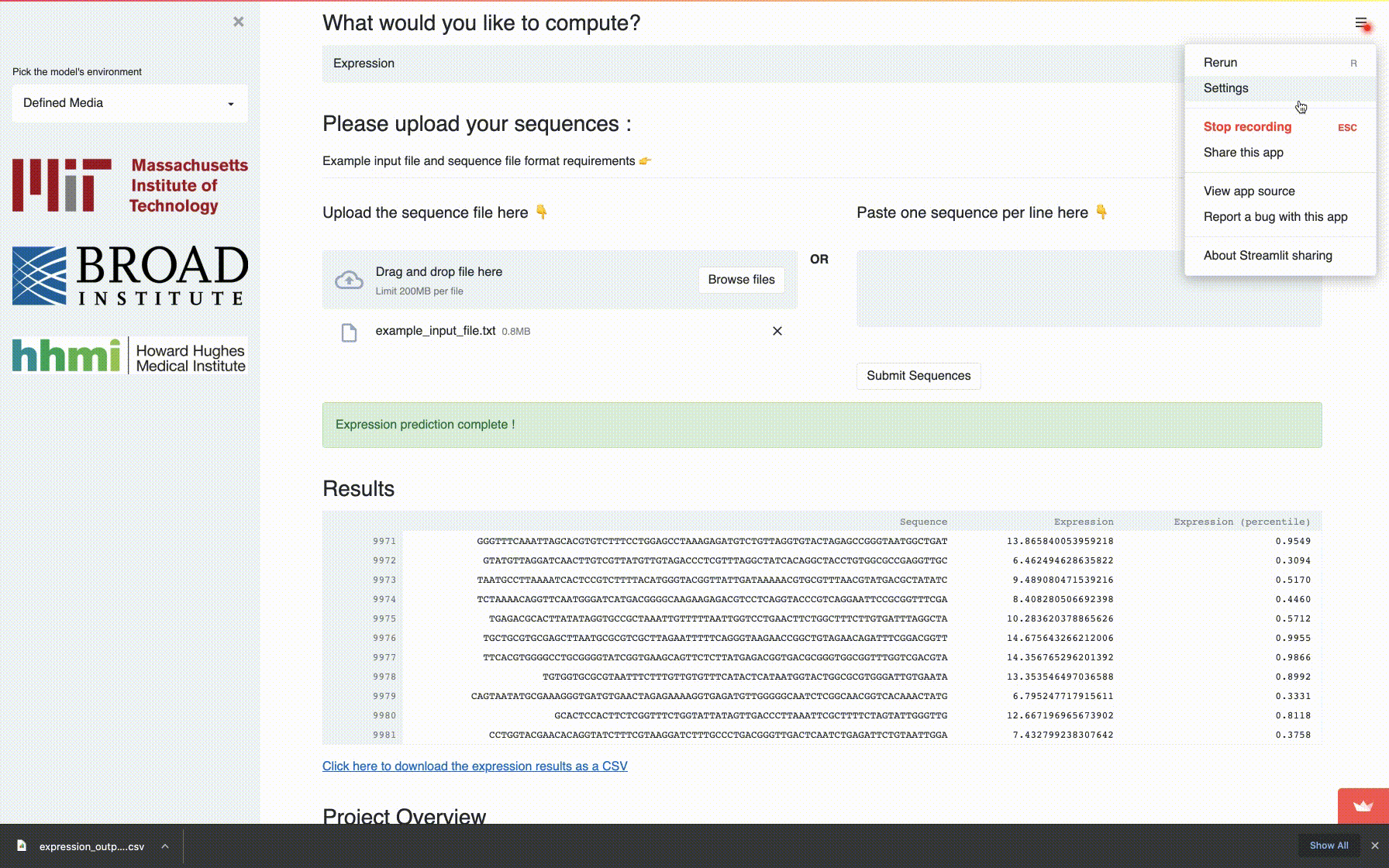
app: A fully self-contained directory for running the interactive application to compute mutational robustness, evolvability vectors and expression. -
manuscript_code: The codebase corresponding to the manuscript. The organization of this directory is further described here.
If you wish to run the app on your local machine or cluster,
- Install Docker.
- Run the following commands on a terminal :
docker pull edv123456789/evolution_app:latest
docker run --rm -d -p 8501:8501/tcp edv123456789/evolution_app:latestThe app is now running and you can access it by navigating to http://localhost:8501/ in your web browser. If running on a remote cluster, you may want to expose port 8501 using ngrok.
- After installing docker and pulling the latest image as described in the first two steps above, run the following on a terminal :
docker run -it --rm --entrypoint /bin/bash edv123456789/evolution_app
python- In the python shell, run :
from app_aux import *
model_condition = 'Glu' #or, 'SC_Ura'
model, _ , __ = load_model(model_condition)
model.summary()You have now loaded our tensorflow.keras model. You may use this as is for downstream computations as described in the manuscript or adapt it for your application (e.g. transfer learning).
To exit the python shell and the docker container, simply press Ctrl+D twice.
Download all data and models used in the manuscript here 👇
DOI : https://doi.org/10.1101/2021.02.17.430503
A comprehensive fitness landscape model reveals the evolutionary history and future evolvability of eukaryotic cis-regulatory DNA sequences, biorXiv 2021.
Eeshit Dhaval Vaishnav*§, Carl G. de Boer*§, Moran Yassour, Jennifer Molinet, Lin Fan, Xian Adiconis, Dawn A. Thompson, Francisco A. Cubillos, Joshua Z. Levin, Aviv Regev§.