Dimensional homogeneity constrained gene expression programming for discovering governing equations from noisy and scarce data
Data-driven discovery of governing equations is of great significance for helping us to understand intrinsic mechanisms and explore physical models. However, it is still not trivial for the state-of-the-art algorithms to discover the unknown governing equations for complex systems. In this work, a novel dimensional homogeneity constrained gene expression programming (DHC-GEP) method is proposed. DHC-GEP discovers the forms of functions and their corresponding coefficients simultaneously, without assuming any candidate functions in advance. Its key advantages, including being robust to the hyperparameters of models, the noise level and the size of datasets, are demonstrated on two benchmarks. Furthermore, DHC-GEP is employed to discover the unknown constitutive relations of two typical non-equilibrium flows. The derived constitutive relations not only are more accurate than the conventional constitutive relations, but also satisfy the Galilean invariance and the second law of thermodynamics. DHC-GEP is a general and promising tool for discovering governing equations from noisy and scarce data in a variety of fields, such as non-equilibrium flows as well as neuroscience, epidemiology, turbulence, and non-Newton fluids.
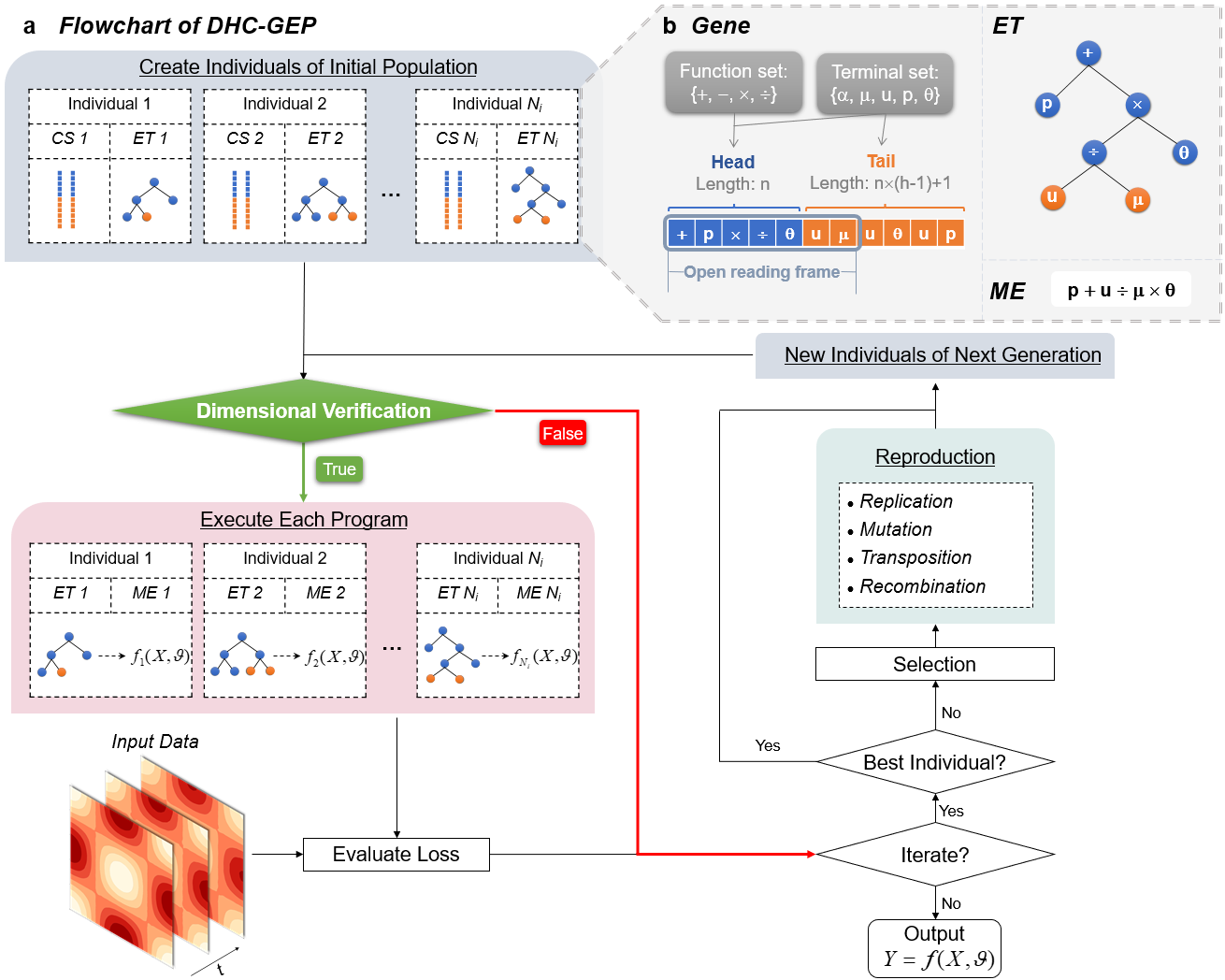
Fig. a is a schematic diagram of DHC-GEP. Initial population is created with
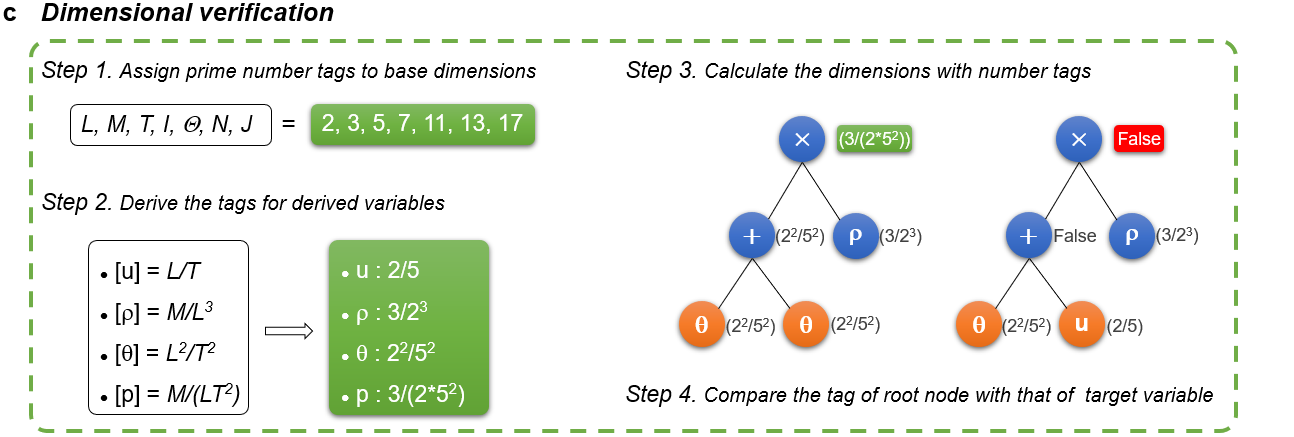
Fig. c shows the strategy of dimensional verification: first assign prime number tags to the base dimensions and derive the tags for the derived variables, then calculate the dimension of each node in the expression tree from the bottom up, finally compare the tag of the root node with that of the target variable. If they are the same, it can be concluded that the certain individual is dimensional homogeneous.
- Python 3.8
- numpy
- geppy
- random
- operator
- pickle
- fractions
- scipy
- time
- tensorflow (1.12.0)
Anoconda is recommended for installing the above dependencies.
All the training data are in the 'data' dictionary.
The scripts are in the corresponding dictionaries. One can run the desired scripts with python.
Every 20 generations, the current optimal individual is checked, and if a new optimal individual appears, it will be output to a '.dat' file in the 'Output' dictionary. The latest population is saved every 20 generations to a '.pkl' file in the 'pkl' dictionary for ease of subsequent restarting if necessary.
If someone wants to employ DHC-GEP in other problems, one should reassign number tags for the imported terminals. This is implemented in the following codes. One can redefine 'dict_of_dimension' as needed. Key is the name of imported terminal. Value is the corresponding number tag.
# Assign prime number tags to base dimensions
L,M,T,I,Theta,N,J = 2,3,5,7,11,13,17
# Derive the tags for dirived physical quantities according to their dimensions
# Note that the tags are always in the form of fractions, instead of floats, which avoids introducing any truncation errors.
# Therefore, we use 'Fraction' function here.
dict_of_dimension = {'rho':Fraction(M,((L)**(3))),
'rho_y':Fraction(M,((L)**(4))),
'rho_yy':Fraction(M,((L)**(5))),
'rho_3y':Fraction(M,((L)**(6))),
'df_c':Fraction((L**2),T)}
# Assign number tags to taget variable
target_dimension = Fraction(M,T*((L)**(3)))