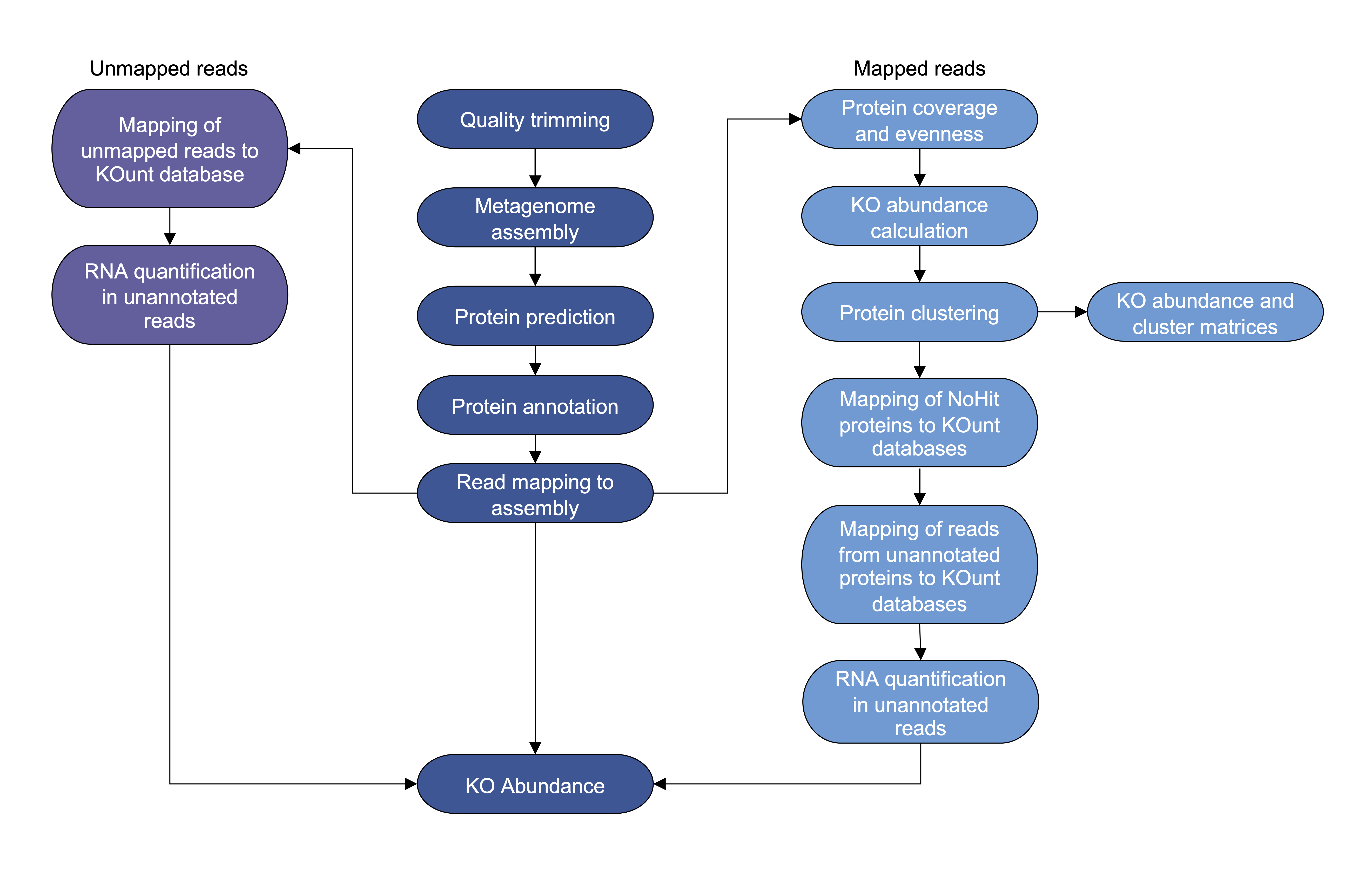
Snakemake pipeline calculating KEGG orthologue abundance in metagenomic sequence data.
KOunt is a Snakemake pipeline that calculates the abundance of KEGG orthologues (KOs) in metagenomic sequence data. KOunt takes raw paired-end reads and quality trims, assembles, predicts proteins and annotates them with KofamScan. The reads are mapped to the assembly and protein coverage calculated. Users have the option of calculating coverage evenness of the proteins and filtering the KofamScan proteins to remove unevenly covered proteins. The proteins annotated by KofamScan are clustered at 100%, 90% and 50% identity within each KO to quantify their diversity; as using the evenness filtering option reduces the numbers of these proteins we don't recommend using the evenness option if you are interested in the clustering results. All predicted proteins that don’t have a KO hit or are excluded by evenness filtering are called 'NoHit’. The NoHit proteins are blasted against a custom UniProt database annotated with a KO and the nucleotides against a custom RNA database. Reads mapped to NoHit proteins that remain unannotated and unmapped reads are blasted against the KOunt databases and RNA quantified in the remaining reads.
If you use KOunt please cite https://academic.oup.com/bioinformatics/article/39/8/btad483/7236497
Download the latest version of the Snakefile, scripts and conda env files.
git clone https://github.com/WatsonLab/KOunt
cd KOunt/
Check that the scripts are executable, if not do: chmod +x scripts/*sh
Download the KOunt UniProt and RNA databases.
wget https://figshare.com/ndownloader/files/37711530
mv 37711530 KOunt_databases.tar
tar -xzvf KOunt_databases.tar
gunzip KOunt_databases_v1/*
rm KOunt_databases.tar
If you wish to update these databases, further information on how they were created is available here.
conda create -n snakemake_mamba -c conda-forge -c bioconda mamba
conda activate snakemake_mamba
mamba install -c bioconda snakemake
conda activate snakemake_mamba
snakemake --use-conda --conda-create-envs-only --cores 1
Download the test fastqs. Leave the raw reads location in the config at default and perform a dry-run with the reads subsampled from ERR2027889. Then run the pipeline. With 8 cores it should take approximately 20 minutes.
wget https://figshare.com/ndownloader/files/39545968
mv 39545968 test_fastqs.tar
tar -xvf test_fastqs.tar
rm test_fastqs.tar
snakemake -k --ri --use-conda -n
snakemake -k --ri --use-conda --cores 8
Amend the options config file, config.yaml, with your fastq file locations and extensions. KOunt expects the raw reads to be in a directory with the same sample name eg. ERR2027889/ERR2027889_R1.fastq.gz. It runs the pipeline on all the samples in the directory you specify in the config file.
To use the default rule all in the Snakefile specify the number of cores you have available and run the entire pipeline with:
snakemake -k --ri --use-conda --cores 8
If you wish to only run part of the pipeline you can specify another rule all.
To perform all steps but the protein clustering use:
snakemake -k --ri --use-conda all_without_clustering --cores 8
To perform all steps but protein clustering and read/protein annotation with the KOunt reference databases:
snakemake -k --ri --use-conda all_without_reference --cores 8
To perform all steps but protein clustering and RNA abundance quantification:
snakemake -k --ri --use-conda all_without_RNA --cores 8
The average run time and maximum memory used by each of the rules on the 10 samples from the KOunt manuscript is available here.
The following options can be amended in the config.yaml file:
raw_readsthe path to the directory containing all the raw reads (default: "reads/")r1_extthe extension of read one (default: "_R1.fastq.gz")r2_extthe extension of read two (default: "_R2.fastq.gz")diamond_dbthe path to the KOunt DIAMOND database (default: "KOunt_databases/KO_DI_1.0.dmnd")mmseq_dbthe path to the KOunt MMseqs2 database (default: "KOunt_databases/KO_RNA_1.0.mmseq")combined_bdgthe path to the KOunt database bedGraph file (default: "KOunt_databases/KO_RNA_DI_1.0.bedgraph")kallistothe path to the KOunt kallisto reference (default: "KOunt_databases/KO_RNA_kallisto_1.0")outdirthe path to the output directory (default: "out/")
The read ids in the trimmed reads are shortened up to the first space and /1 or /2 added to the end if not already present. By default the read ids are compared to ensure all ids are unique but this can be changed if you're sure they will be
checking_fqsrunning fastq_utils to check that the read ids are unique and the fastq is valid (default: ""). Change to "#" if not checkingnot_checking_fqsnot running fastq_utils on the reads (default: "#"). Change to "" if checking
The abundance of proteins that KofamScan annotates with multiple KOs can either be split between the KOs or summed with all other proteins with multiple hits into 'Multiples'
splitting_multiplessplitting proteins between their KO hits (default: ""). Change to "#" if grouping multiplesgrouping_multiplesgrouping proteins that have multiple KO hits (default: "#"). Change to "" if grouping multiples
r1adthe adapter sequence for read one (default: "AGATCGGAAGAGC")r2adthe adapter sequence for read two (default: "AGATCGGAAGAGC")polyguse -g to enable trimming of polyG tails (default: "")qualuse -Q to disable quality filtering (default: "")minlenthe minimum required read length (default: "50")overlapthe minimum required length of an overlap of PE reads (default: "5")trim_threadsthe number of threads (default: "4")
mega_memthe maximum memory megahit can use (default: "4.8e+10”)mega_threadsthe number of threads (default: "8")mega_kmersthe kmer sizes (default: "27,37,47,57,67,77,87")mega_lenthe minimum required contig length (default: "300")
bwa_threadsthe number of threads (default: "4")
cov_splitthe number of chunks to split the BAM file into. The bigger the number of chunks, the memory required decreases but run time increases (default: “10”)evenness_yesif you want to filter the kofamscan results by coverage evenness then leave this option empty (default: "")evenness_noif you don't want to filter the kofamscan results by coverage evenness then leave this option empty. Must be the opposite of evenness_yes (default: "#")
dbthe path to the kofamscan database. Amend if you have an alternate version you wish to use, default will download the current database version (default: “out/Kofamscan/kofam/”)
kofamscan_threadsthe number of threads (default: “4”)
evenness_pidthe evenness percentage threshold to filter the proteins by (default: "0.95")
cdhit_memthe maximum memory CD-HIT can use (default: “32000”)cdhit_threadsthe number of threads (default: “8”)
mmseq_keggs_threadsthe number of threads (default: “8”)
mmseq_nohit_threadsthe number of threads (default: “8”)
dia_threadsthe number of threads (default: “8”)min_qcthe minimum percentage of the length of the reference hit that the protein has to be (default: “90”)max_qcthe maximum percentage of the length of the reference hit that the protein has to be (default: “110”)min_pidthe minimum percentage identity required to be classed as a hit (default: “80”)
barrnapthe number of threads (default: “4”)
nohitthe number of threads (default: “8”)
kallisto_threadsthe number of threads (default: “8”)
unmapped_threadsthe number of threads (default: “8”)
Results/KOunts_Kofamscan.csv KO abundance in each sample, calculated by Kofamscan, without read mapping
Results/All_KOunts_nohit_unmapped_default.csv Final KO abundance in each sample
Results/Number_of_clusters.csv Number of clusters of proteins at 90% and 50% sequence identity in each KO, the number of clusters that contain multiple proteins and the number of singleton clusters
Results/KOunts_Kofamscan.csv KO abundance in each sample, calculated by Kofamscan, without read mapping
Results/All_KOunts_nohit_unmapped_no_clustering.csv Final KO abundance in each sample
Results/All_KOunts_without_reference.csv Final KO abundance in each sample calculated by Kofamscan, without read mapping
Results/KOunts_Kofamscan_without_clustering.csv KO abundance in each sample, calculated by Kofamscan, without read mapping
Results/All_KOunts_without_RNA.csv Final KO abundance in each sample