DocGenome: An Open Large-scale Scientific Document Benchmark for Training and Testing Multi-modal Large Language Models
Scientific documents record research findings and valuable human knowledge, comprising a vast corpus of high-quality data. Thus, leveraging multi-modality data extracted from these documents and assessing large models' abilities to handle scientific document-oriented tasks is meaningful. Despite promising advancements, large models still perform poorly on multi-page scientific document extraction and understanding tasks, and their capacity to process within-document data formats such as charts and equations remains under-explored. To address these issues, we present DocGenome, a structured document dataset constructed by annotating 500K scientific documents from 153 disciplines in the arXiv open-access community, using our custom auto-labeling pipeline. DocGenome features four characteristics:
-
- Completeness: It is the first dataset to structure data from all modalities including 13 layout attributes along with their \LaTeX\ source codes.
-
- Logicality: It provides 6 logical relationships between different entities within each scientific document.
-
- Diversity: It covers various document-oriented tasks, including document classification, visual grounding, document layout detection, document transformation, open-ended single-page QA and multi-page QA.
-
- Correctness: It undergoes rigorous quality control checks conducted by a specialized team.
Besides, based on DocGenome, we conduct extensive experiments to demonstrate the advantages of DocGenome and objectively evaluate the performance of current large models on our benchmark.
- [2024/7/23] We have supported TestSet downloads from Huggingface. If you want to evaluate your model on TestSet, please refer to Evaluation.
- [2024/7/12]] We have supported dataset downloads from Huggingface.
- [2024/6/15] 🔥 Our paper entitled "DocGenome: An Open Large-scale Scientific Document Benchmark for Training and Testing Multi-modal Large Models" has been released in arXiv Link
- [2024/6/6] 🔥 We have released the DocGenome benchmark, includes 8 subsets as follows:
| Datasets | # Discipline | # Category of Units | # Pages in Train-set | # Pages in Test-set | # Task | # Used Metric | Publication | Entity Relations |
|---|---|---|---|---|---|---|---|---|
| DocVQA | - | N/A | 11K | 1K | 1 | 2 | 1960-2000 | ❎ |
| DocLayNet | - | 11 | 80K | 8K | 1 | 1 | - | ❎ |
| DocBank | - | 13 | 0.45M | 50K | 3 | 1 | 2014-2018 | ❎ |
| PubLayNet | - | 5 | 0.34M | 12K | 1 | 1 | - | ❎ |
| VRDU | - | 10 | 7K | 3K | 3 | 1 | - | ❎ |
| DUDE | - | N/A | 20K | 6K | 3 | 3 | 1860-2022 | ❎ |
| D^4LA | - | 27 | 8K | 2K | 1 | 3 | - | ❎ |
| Fox Benchmark | - | 5 | N/A (No train-set) | 0.2K | 3 | 5 | - | ❎ |
| ArXivCap | 32 | N/A | 6.4M* | N/A | 4 | 3 | - | ❎ |
| DocGenome (ours) | 153 | 13 | 6.8M | 9K | 7 | 7 | 2007-2022 | ✅ |
We provide 8 subsets of DocGenome-train for downloading:
Data Download
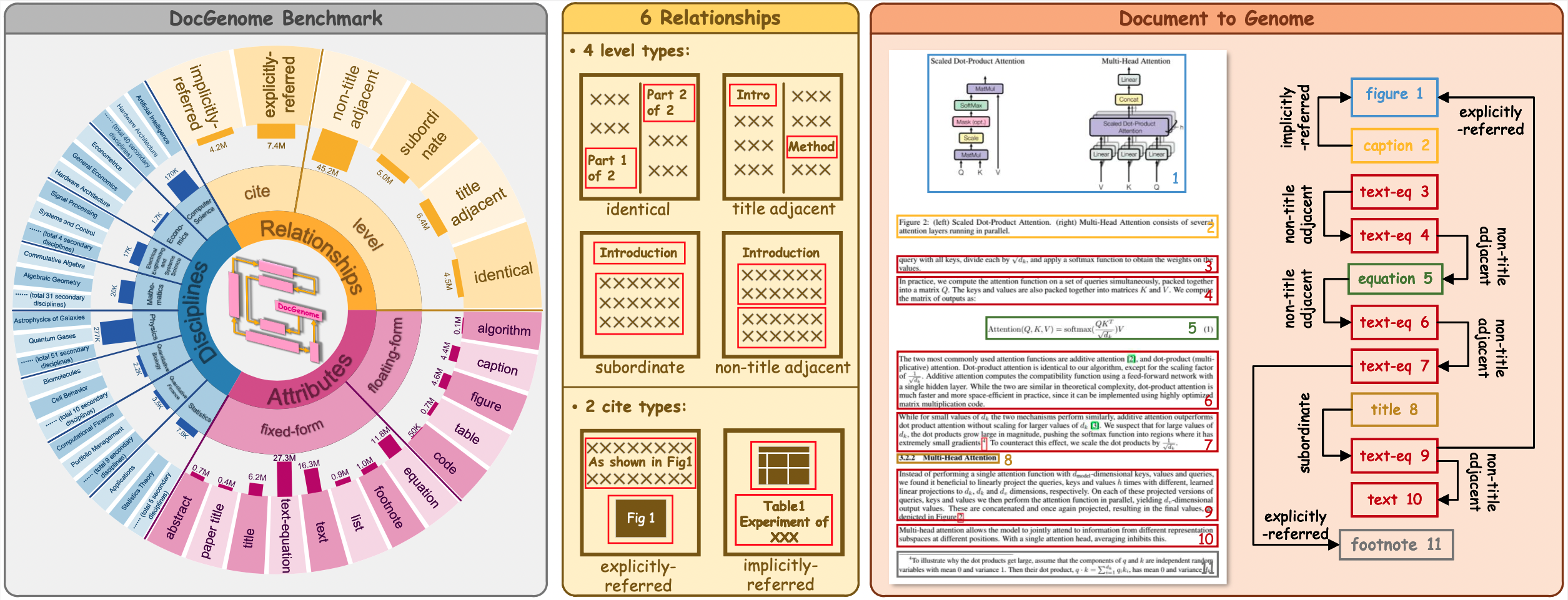
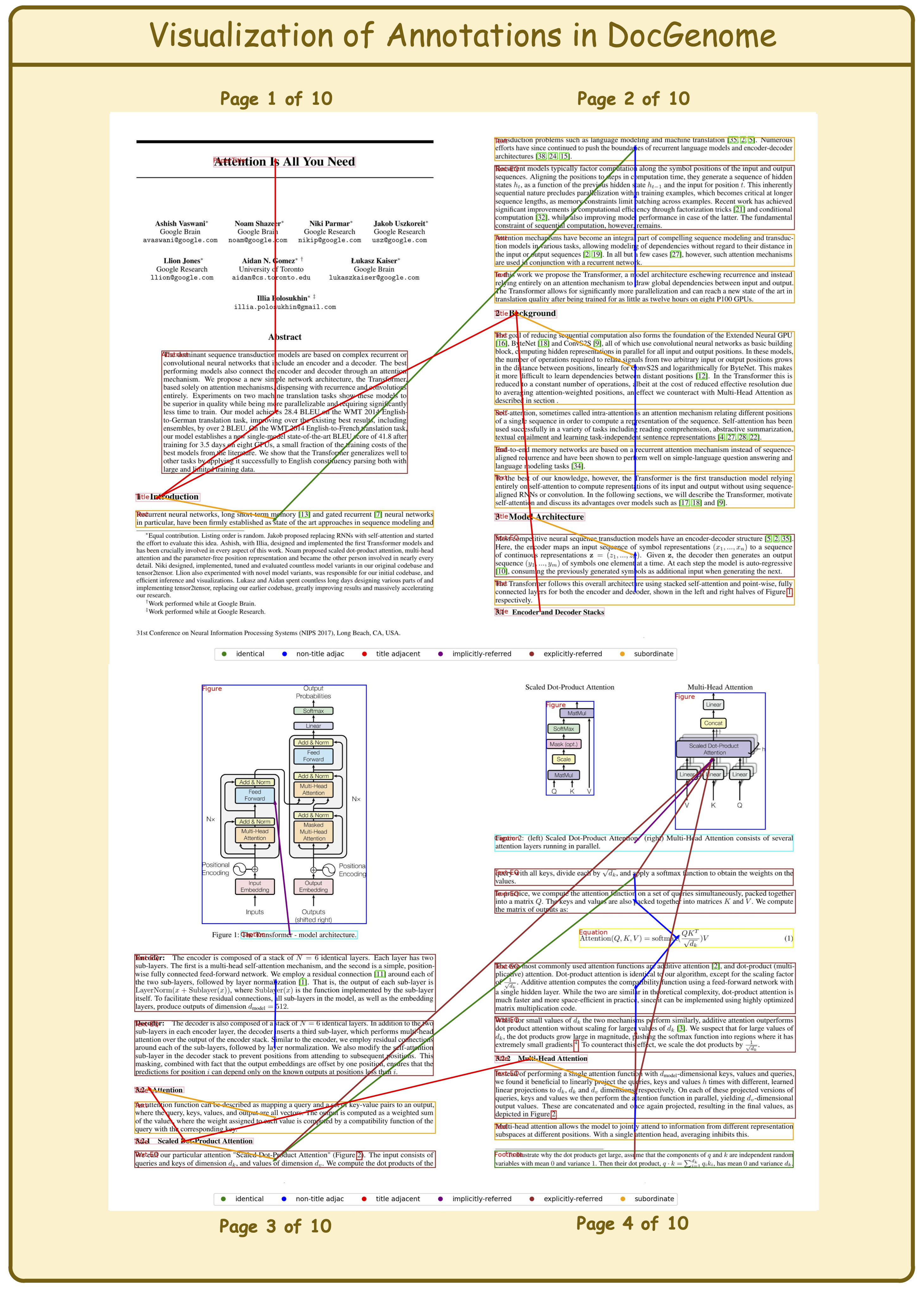
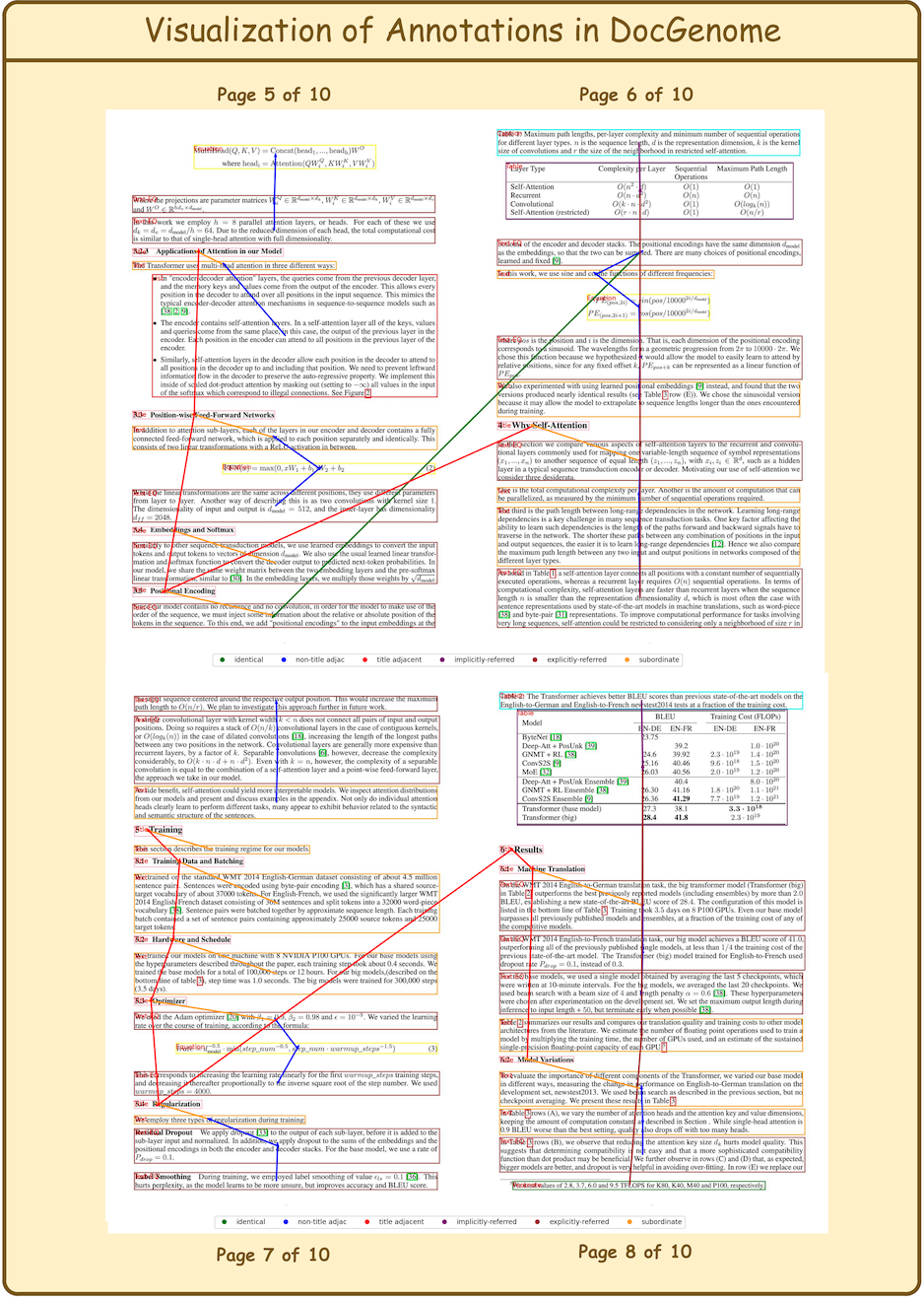
DocGenome contains 4 level relation types and 2 cite relation types, as shown in the following table:
| Name | Description | Example |
|---|---|---|
| Identical | Two blocks share the same source code. | Cross-column text; Cross-page text. |
| Title adjacent | The two titles are adjacent. | (\section{introduction}, \section{method}) |
| Subordinate | One block is a subclass of another block. | (\section{introduction}, paragraph within Introduction) |
| Non-title adjacent | The two text or equation blocks are adjacent. | (Paragraph 1, Paragraph 2) |
| Explicitly-referred | One block refers to another block via footnote, reference, etc. | (As shown in \ref{Fig: 5} ..., Figure 5) |
| Implicitly-referred | The caption block refers to the corresponding float environment. | (Table Caption 1, Table 1) |
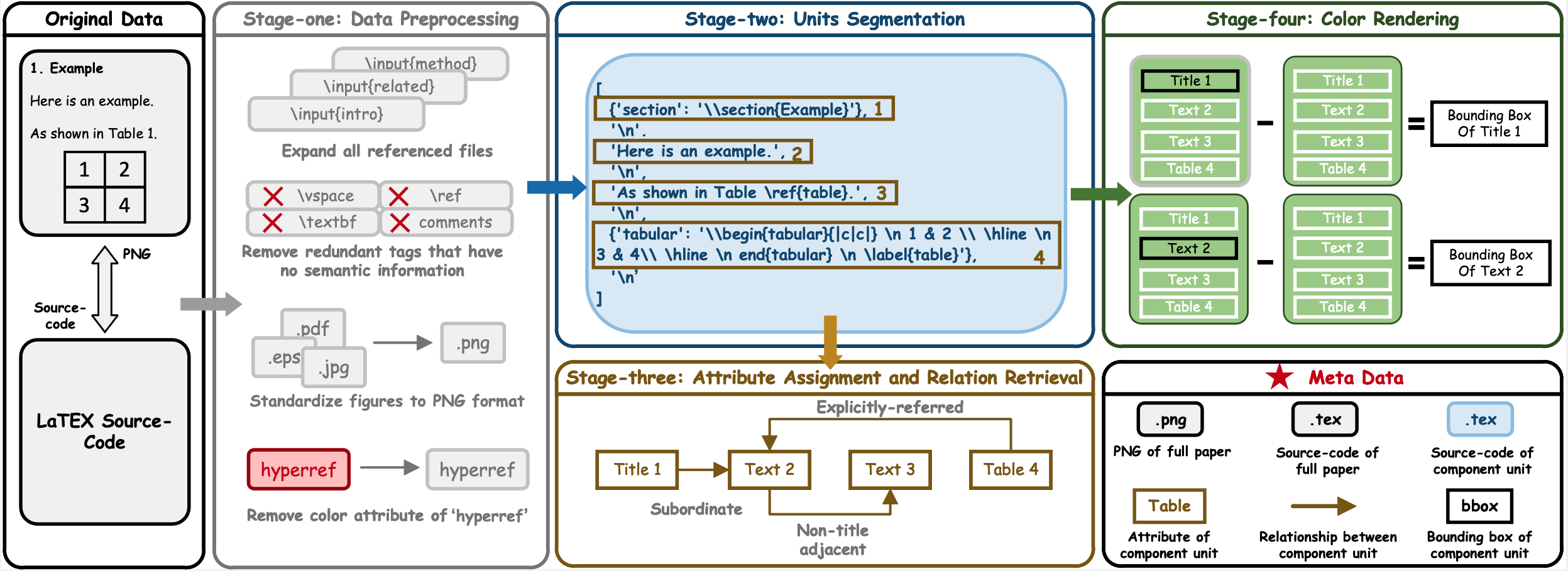
DocGenome has 13 attributes of component units, which can be categorized into two classes
- 1) Fixed-form units, including Text, Title, Abstract, etc., which are characterized by sequential reading and hierarchical relationships readily discernible from the list obtained in Stage-two of the designed DocParser.
- 2) Floating-form units, including Table, Figure, etc., which establish directional references to fixed-form units through commands like \texttt{\textbackslash ref} and \texttt{\textbackslash label}.
| Index | Category | Notes |
|---|---|---|
| 0 | Algorithm | |
| 1 | Caption | Titles of Images, Tables, and Algorithms |
| 2 | Equation | |
| 3 | Figure | |
| 4 | Footnote | |
| 5 | List | |
| 7 | Table | |
| 8 | Text | |
| 9 | Text-EQ | Text block with inline equations |
| 10 | Title | Section titles |
| 12 | PaperTitle | |
| 13 | Code | |
| 14 | Abstract |
Page distribution of DocGenome. 20% of documents are five pages or fewer, 50% are ten pages or fewer, and 80% are nineteen pages or fewer.
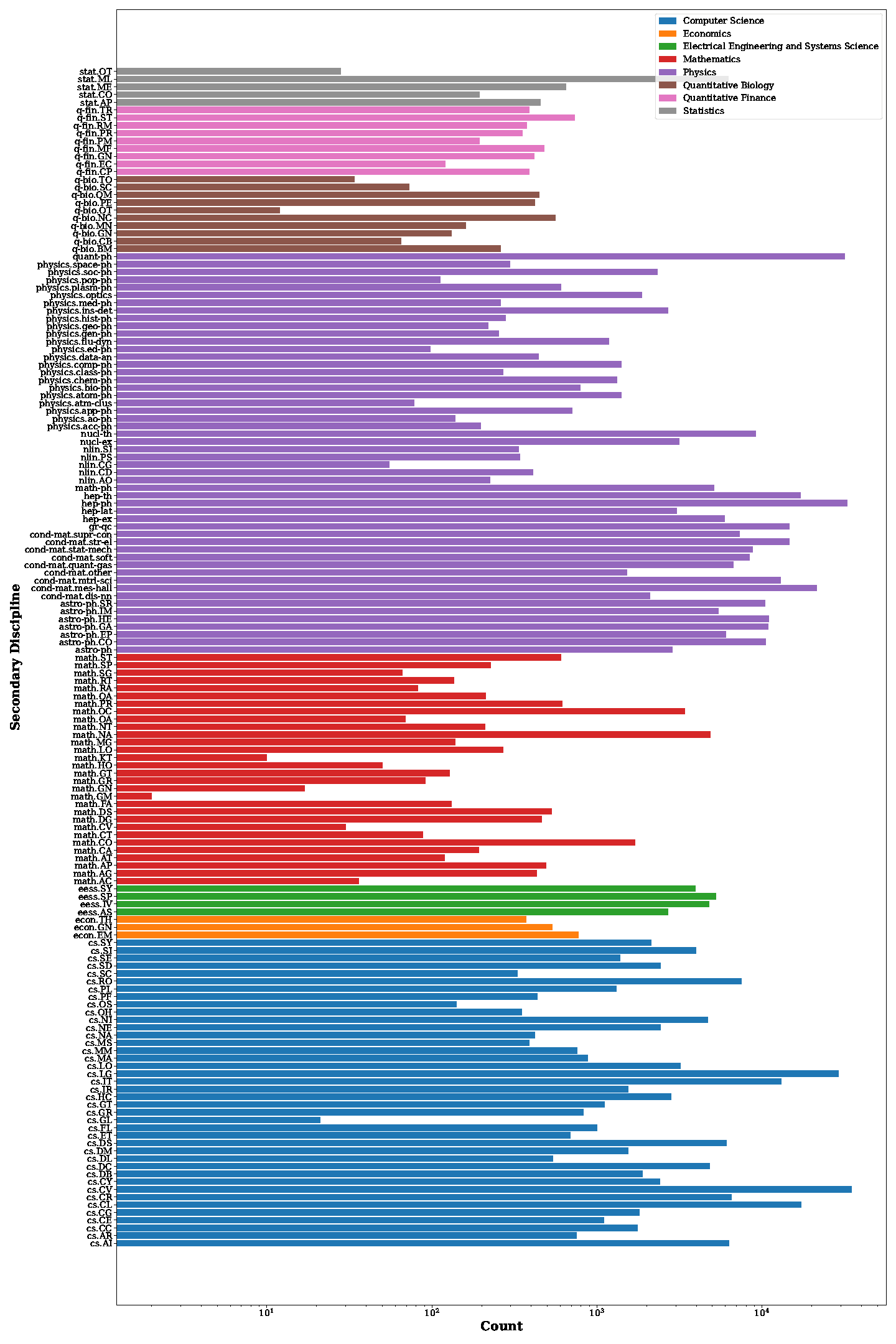
Distribution of secondary disciplines in our DocGenome. The count on the x-axis represents the number of documents, and documents from the same primary discipline are marked with the same color.
If you find our work useful in your research, please consider citing Fox:
@article{xia2024docgenome,
title={DocGenome: An Open Large-scale Scientific Document Benchmark for Training and Testing Multi-modal Large Language Models},
author={Xia, Renqiu and Mao, Song and Yan, Xiangchao and Zhou, Hongbin and Zhang, Bo and Peng, Haoyang and Pi, Jiahao and Fu, Daocheng and Wu, Wenjie and Ye, Hancheng and others},
journal={arXiv preprint arXiv:2406.11633},
year={2024}
}