- Install the latest version of R (4.0.2)
- Install
CERINAdependencies from CRAN:
install.packages(c("shiny", "shinydashboard", "shinyjs", "shinycssloaders", "shinyBS", "DT", "tidyverse",
"dendextend", "visNetwork", "heatmaply", "Matrix", "fastcluster", "htmltools", "reshape2", "igraph"))CERINAcan be run with the following command:
shiny::runGitHub("jcardenas14/CERINA")The CERINA application was developed as a resource for researchers to infer circRNA function under the competing endogenous RNA (ceRNA) framework outlined in Cerina paper. Paired circRNA, linear RNA, and miRNA expression data across 11 human tissues from ENCODE were analyzed and integrated using Pareto optimality to rank circRNA-miRNA and miRNA-linRNA interactions. These rankings are the driving force behind the Cerina methodology and many of the visualizations/tables users will encounter in the app. All data has been processed, filtered, and normalized according to the procedures outlined in the paper.
By default, linear RNA and miRNA data are reported using CPM (counts per million) and circRNA data are reported using SRPBM (spliced reads per billion mapping). However, users are given the option to visualize all figures/tables using DESeq2 normalized counts via check-boxes. Throughout the documentation, we refer to normalized counts (either CPM, SRPBM, or DESeq2) as expression.
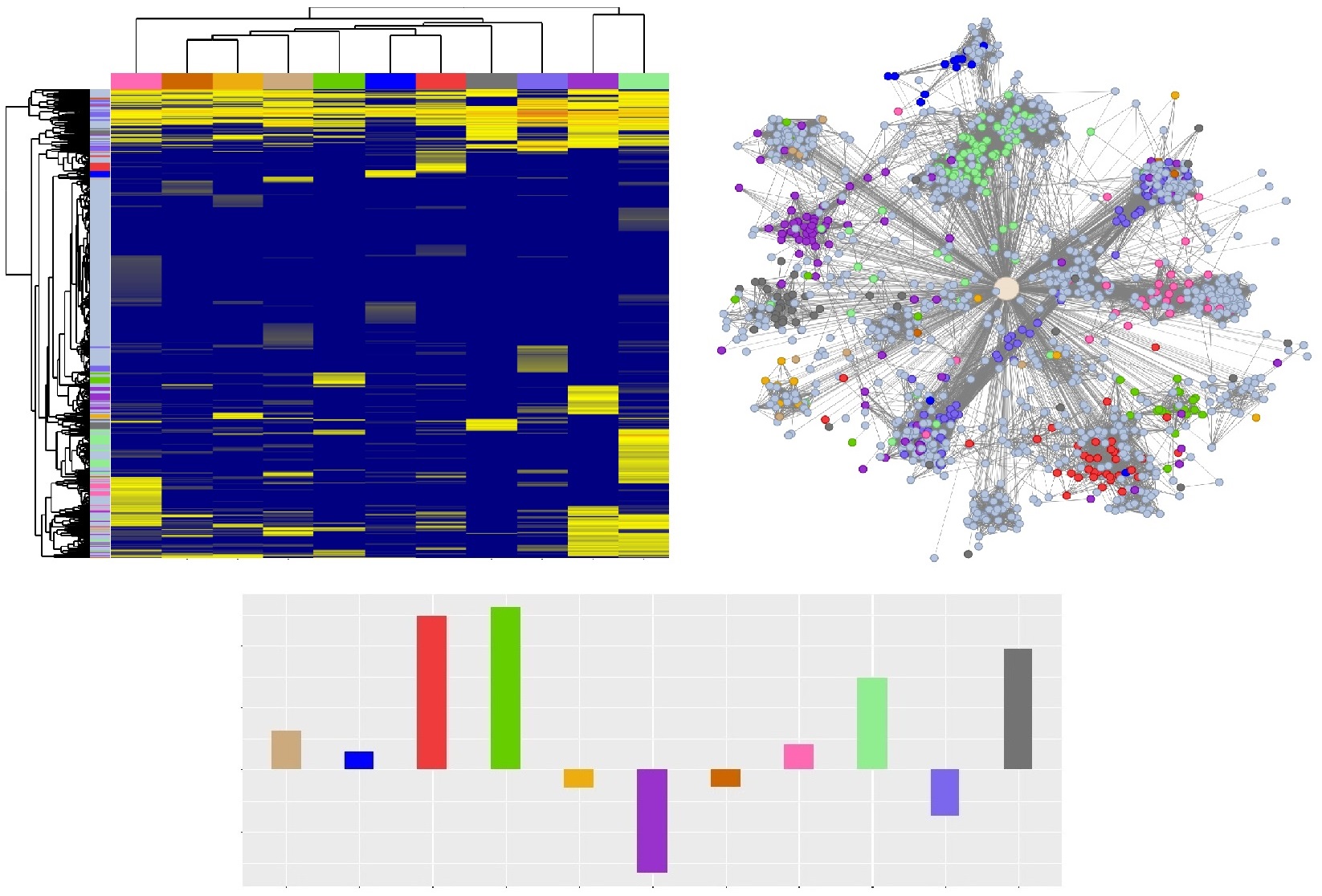
We defined a circRNA as "tissue specific" if, for a given tissue, it has mean log2 DESeq2 normalized count ≥ 2 and at least 2 fold higher expression compared to the second highest expressed tissue. Throughout CERINA, circRNAs are colored according to their tissue specificity and are labeled as "unspecific" if the criteria are not met.
Numerous interactions are depicted in CERINA via networks whose edges have been weighted to reflect the strength of interaction:
- circRNA-miRNA - weighted by pareto interaction score
- miRNA-gene - weighted by pareto interaction score
- circRNA-circRNA - weighted by spearman correlation
- circRNA-gene - weighted by empircal CDF of -log10(pvalue), where pvalues are obtained from the gene level permutation test.
All other interactions (i.e. gene-function, miRNA-function, circRNA-function) have been given the same weight of 1.
-
linear RNA - Search by Ensembl ID or gene symbol.
-
miRNA - Search using 'hsa-xxx-xxx' format.
-
circRNA - Search using chromosome location, ciri2 ID, circBase ID, circAtlas ID (hg19), or parental gene symbol. When searching by gene symbol, an additional input appears listing all circRNAs sharing the same parental gene; users can then select a circRNA from the list.