Updated on 2023.06.20
This repository provides the official implementation of the Ultrasound foundation model (USFM) for ultrasound image downstream tasks.
key feature bulletin points here:
- The model was pre-trained on over 2M ultrasound images from five different tissues.
- We used a pre-training strategy based on masked image modeling (BEiT) with more sensitivity to structure and texture.
- The pre-trained model achieves SOTA performance on multiple ultrasound image downstream tasks. A more extensive test is in progress
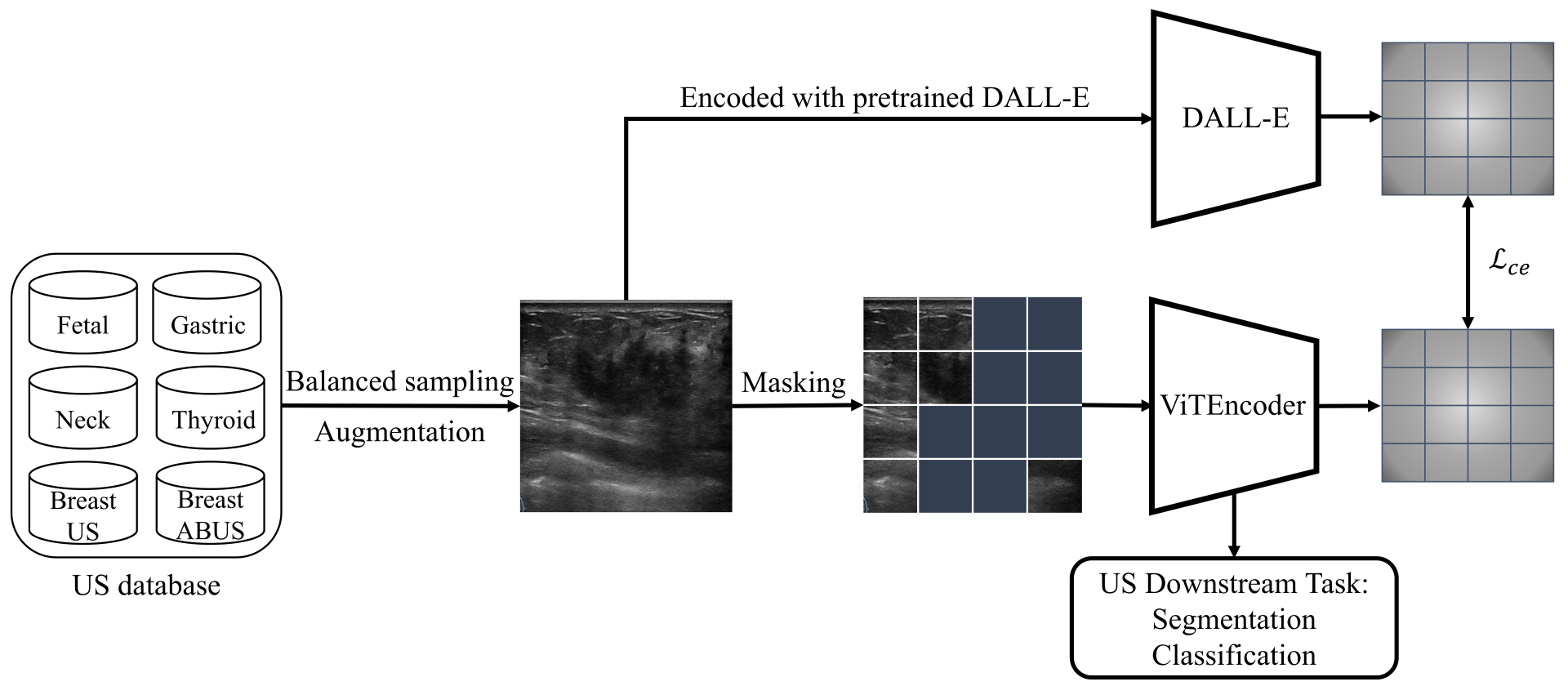
Our ultrasound foundation model (USFM) is pre-trained on the database containing ultrasound images of six different tissues. The most popular encoder, visual transformer (ViT), was chosen as the base architecture. For the pre-training strategy, we refer to BEiT and use the fully trained DALL-E as a strong Teacher to guide our model to learn the proper feature representation. Experimental results demonstrate that our model has excellent performance on ultrasound image downstream tasks.
- Pip
# clone project
git clone https://github.com/openmedlab/USFM.git
cd USFM
# [OPTIONAL] create conda environment
conda create -n USFM python=3.9
conda activate USFM
# install pytorch according to instructions
# https://pytorch.org/get-started/
# install requirements
pip install -r requirements.txt- Conda
# clone project
git clone https://github.com/openmedlab/USFM.git
cd USFM
# create conda environment and install dependencies
conda env create -f environment.yaml
# activate conda environment
conda activate USFM
pip install -U openmim
mim install mmcv# In the folder USFM
pip install -v -e .USFM is pre-trained on 3 private and 4 public datasets using BEIT under the manner of feature reconstruction. Several datasets were collected as downstream tasks for validation. Here, we provide 2 public datasets for the ultrasound downstream task.
- tn3k [link: https://drive.google.com/file/d/1jPAjMqFXR_lRdZ5D2men9Ix9L65We_aO/view?usp=sharing\]
- tnscui [link: https://drive.google.com/file/d/1Ho-PzLlcceRFdu0Cotxqdt4bXEsiK3qA/view?usp=sharing\]
# mkdir data/Download the dataset from Google Drive tn3k and tn3k and save it in folder data.
# set the Dataset name (one of tn3k, tnscui)
export dataset=tn3k
# unzip dataset
tar -xzf $dataset.tar.gz $dataset/Download the model weight from Google Drive USFMpretrained and save it in folder assets as USFMpretrained.ckpt.
python usfm/train.py tag=seg_$dataset experiment=ftSeg.yaml model.net.backbone.pretrained=assets/USFMpretrained.ckpt data=$dataset data="{batch_size:40, num_workers:4}" trainer="{devices:[0,1], strategy:ddp}"The fine-tuning segmentation results for publicly dataset [TN3K and tnscui] are shown in the table.
| Dataset | Model | Architecture | Dice |
|---|---|---|---|
| TN3K | non-pretrained | UPerNet(ViT-B) | 0.860 |
| TN3K | SAM-encoder | - | 0.818 |
| TN3K | USFM | UPerNet(ViT-B) | 0.871 |
| tnscui | non-pretrained | UPerNet(ViT-B) | 0.879 |
| tnscui | SAM | - | 0.860 |
| tnscui | USFM | UPerNet(ViT-B) | 0.900 |
- Webpage
- Social media
This project is under the CC-BY-NC 4.0 license. See LICENSE for details.
Our code is based on BEiT, transformer, pytorch-image-models , and lightning-hydra-template . Thanks them for releasing their codes.
Have a question? Found a bug? Missing a specific feature? Feel free to file a new issue, discussion or PR with respective title and description.
Please perform a code check before committing with the pre-commit hooks.
# pip install pre-commit
pre-commit run -aUpdate pre-commit hook versions in .pre-commit-config.yaml with:
pre-commit autoupdate