``This repository provides a PyTorch version implementation of the paper "3D Registration of pre-surgical Prostate MRI and Histopathology Images via super-resolution volume reconstruction" (Link).
This paper utilizes two public prostate datasets and an internal dataset from Stanford Hospital. Therefore we only provide the link to the public datasets, links are shown below:
Following the paper, we only take a subset with all T2w-related volumes to train the model.
This code is implemented on Ubuntu 20.04, the command may need slight changes to fit Windows users.
- Ubuntu 20.04
- Python 3.9
- PyToch 1.11
- CUDA 11.2
Please use pip install -r requirements.txt to install the required packages in advance.
-
Remanage dataset
- We regenerate
.pklfiles to save the volumes before starting training. - You should follow the Python script in
./revise_datasetsdir to generate the datasets on your device. Pay attention to the outputmeta_data_train.jsonand themeta_data_test.jsonfile, which is important for Dataloader during the coming training.
- We regenerate
-
Then you can start the multi-GPU training with the following commands:
torchrun \
--standalone \
--nnodes=1 \
--nproc_per_node=2 \
train_main.py \
--world_size 2 \
--data_path "/root/Dataset/prostate" \
--meta_data_path /root/Dataset/meta_data_train.json \
--batch_size 4 \
--epochs 81 \
--update_d_period 5 \
--lr 1e-4 \
--weight_decay 0.05 \
--warmup_epochs 2 \
--output_dir ./output_dir/ \
--log_dir ./output_dir/ \
2> train_error.log 1>train_logs.log
You need to change the argument data_path to the path containing your generated .pkl file, and the argument meta_data_path to the meta_data_train.json file path.
Note that if you change the training datasets, you may have to re-balance the
update_d_periodparametes to make sure the discriminator and the generator still balanced well. Besides, it may need lessepochdepends on your task.
torchrun \
--standalone \
--nnodes=1 \
--nproc_per_node=2 \
evaluate_main.py \
--world_size 2 \
--data_path /root/Dataset/prostate \
--meta_data_path /root/Dataset/meta_data_test.json \
--checkpoint_path /root/workspace/srGAN/output_dir \
--batch_size 32 \
--epochs 70 \
--output_dir ./eval_dir/ \
--log_dir ./eval_dir/ \
2> test_error.log 1>test_logs.log
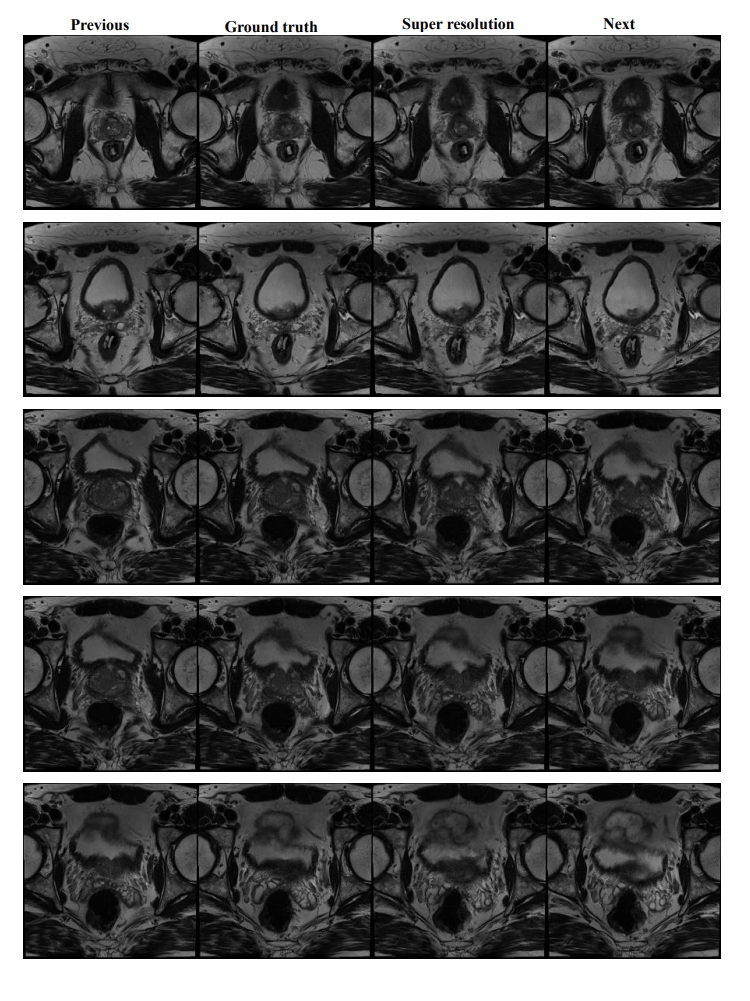
After running this code, you will find the output results under your --output_dir. It should look like this:
@article{
Sood_Shao_Kunder_Teslovich_Wang_Soerensen_Madhuripan_Jawahar_Brooks_Ghanouni_et al._2021,
title={3D Registration of pre-surgical prostate MRI and histopathology images via super-resolution volume reconstruction},
volume={69},
ISSN={13618415},
DOI={10.1016/j.media.2021.101957},
journal={Medical Image Analysis},
author={Sood, Rewa R. and Shao, Wei and Kunder, Christian and Teslovich, Nikola C. and Wang, Jeffrey B. and Soerensen, Simon J.C. and Madhuripan, Nikhil and Jawahar, Anugayathri and Brooks, James D. and Ghanouni, Pejman and Fan, Richard E. and Sonn, Geoffrey A. and Rusu, Mirabela},
year={2021},
month={Apr},
pages={101957},
language={en}
}