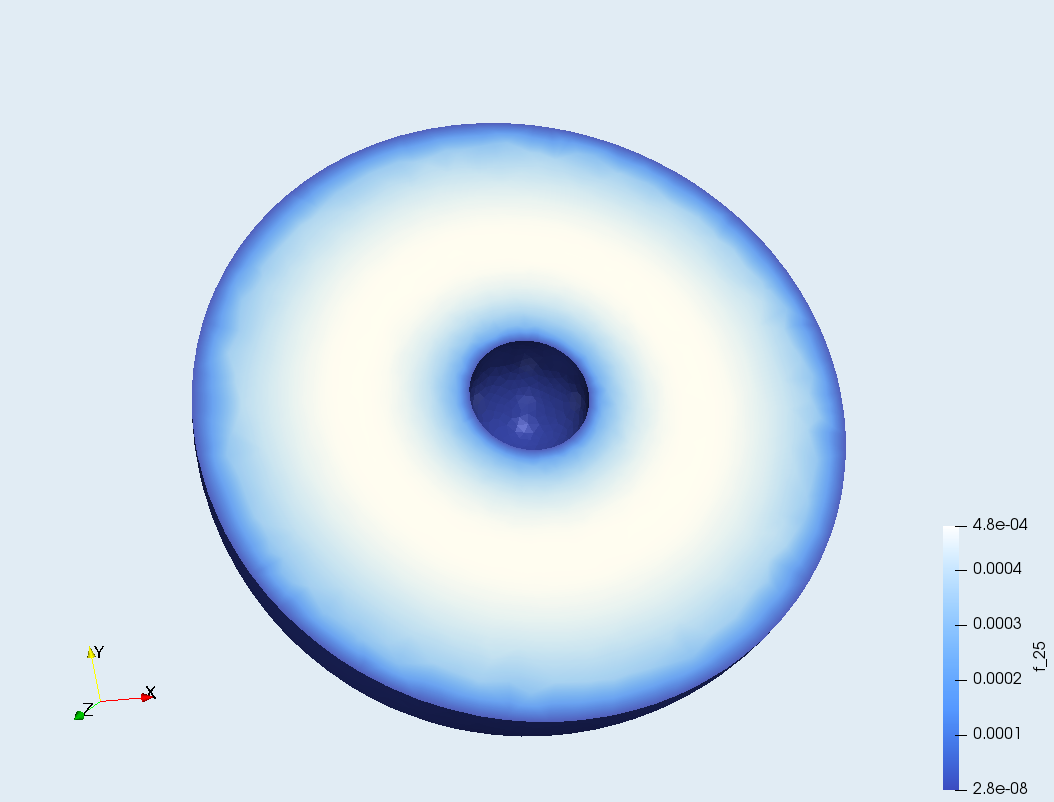
3D finite element analysis of Chameleon field
codes & documentations both under development
-
Fenics Project
This project 100% relies on this pde solver core package. -
Fenics Tutorial
Not the quickest way to learn, but very long and in-depth tutorial with lots of information. -
Fenics 2D Linear Poisson example
Note that there are multiple versions of documentations available online. Currently Dolfin (interface module for Fenics) 1.6.0 is the newest version in March 2020. The version of the documentation can be checked with the version number printed on the left-top corner. -
Blog article about Electrostatic sim with Fenics
Best blog article for the beginner of Fenics 3D simulation. I initially built a copy of this sample code, and modified parts by parts to learn how it works. -
Fenics online forum
Very active. Quickest way to get your question answered as long as you follow the rule. -
Variational Formulation of elliptic PDEs
Technical, but good for learning. Just reading the first few pages is a good way to start learning about this technique.
- Chameleon Dark Energy and Atom Interferometry (Elder et al. 2016)
- Atom-interferometry constraints on dark energy (Hamilton et al. 2015)
Fenics is an open-source pde solver mainly developed for the finite element analysis. It runs c++ code wrapped by python package. The documentation is sometimes outdated and confusing, but the online forum is very active and main developers (researchers at Cambridge University) typically answer to any reasonable question within a day.
In this test example, we model the vacuum space between vacuum chamber and the test source. We set up the boundary condition at the surface. This is only valid for a very strong coupling (=the field reaches the equilibrium very quickly near the surface of heavy objects), and the full solution to the non-approximate model is presented in a separate notebook.
- open FreeCAD (
$freecad) - make 3D model
- subtract objects to create single volume (in this example)
- save as
filename.step
- open GMSH (
$gmsh) - Geometry -> Physical groups -> add -> Surface/Volume: Assign physical groups (surfaces and a volume) for each boundary : make sure the assigned number is the index used for the boundary condition. In this case we assign surface #0 for outer sphere, surface #1 for inner sphere, and volume #3 for the volume (we only have one volume).
- Mesh -> Define -> Transfinite -> Line: Define the number of vertex points along the line that defines the surface of sphere (blue curve).
- Mesh -> 3D to generate mesh
- save as
filename.msh
$dolfin-convert filename.msh filename.xml: This will create three files (*.xml,*_physical_region.xml, and*_facet_region.xml).
To perform the simulation, we need to:
- (1) Define necessary parameters with proper scaling of units
- (2) Set up boundary conditions
- (3) Define pde to be solved
- (4) Run
- (5) Plot & check results.
- surfaces: constant equilibrium value (
$\sqrt{\Lambda^5M/\rho}$ ) where$\nabla^2\phi=0$ for a thick wall (e.g. vacuum chamber)
- Write down the poisson equation to be solved
- Rewrite the equation in the variational form
- Rescale
The Chameleon Field is expressed (in non-relativistic case) $$ \nabla^2\phi = V_{\mathrm{eff},\phi}$$ where $$ V_{\mathrm{eff}} = V(\phi) + A(\phi)\rho_m$$ and $$ \begin{cases} V(\phi) = \Lambda^4\left(1 + \frac{\Lambda^n}{\phi^n}\right)\ A(\phi) \approx 1 + \frac{\phi}{M}\end{cases}$$ We take n=1. Then $$ V_{\mathrm{eff}} = \Lambda^4\left(1 + \frac{\Lambda}{\phi}\right) + \left(1 + \frac{\phi}{M}\right)\rho_m$$
So the equation to be solved is $$ -\nabla^2\phi + \frac{\rho_m}{M} = \frac{\Lambda^5}{\phi^2}$$
Using the test function
$$ F \equiv \int\left(\nabla\phi \cdot \nabla v + \frac{\rho_m}{M} v\right)\mathrm dx
- \left(\int\frac{\Lambda^5}{\phi^2}v\mathrm dx + \int gv\mathrm ds\right) = 0$$
Here
Note that this equation must be evaluated in natural units. We use 1eV as the unit scale.
In the pde solver, however, the unit of length is [mm] (or any unit the 3D mesh is initially modeled in.
To rescale this equation into the proper unit, we introduce the scaling constant
where
This gives the rescaled Poisson equation
Since the units of
Thus, with
$$ F \equiv \int\left(\lambda^2\tilde\nabla\phi \cdot \tilde\nabla v + \frac{\rho_m}{M} v\right)\lambda^{-1}\mathrm d\tilde x
- \left(\int\frac{\Lambda^5}{\phi^2}v\lambda^{-1} \mathrm d\tilde x + \int gv\mathrm \lambda^{-1} d\tilde s\right) = 0$$
is the final form of the pde for Chameleon field.
There was a significant change in the mesh importing method. With some community help at Fenics forum, now we have a basic set of functions to import mesh.
references:
https://fenicsproject.discourse.group/t/subdomain-for-different-materials-based-on-physical-volume/3541/
https://fenicsproject.discourse.group/t/multiple-domains-with-gmsh-any-tutorial-available/
@dokken Thank you for the suggestion. I have solved my issue, but for other >people who might be looking for a tip (that are not trivial to beginners), I will >describe my issue and the solution below:
After hours of investigation, I have noticed there were three main issues I've >had:
My model is consisted of two objects, and each of them are stored as different >elements in
msh.cellsarray. This means there are two separated cell data >sections whose type is 'tetra' (and same goes to 'triangle'), and simply doingfor cell in msh.cells: if cell.type == "tetra": tetra_cells = cell.data elif cell.type == "triangle": triangle_cells = cell.datawill fail to include all of such cell data because this code overwrites every >time it encounters a new cell data array. To fix this, I changed the code to >cocatenate those data. Below is a minimum working example of my mesh convert >function. I am sure there is more elegant way of doing this, but at least this >code works.
def convertMesh(mshfile): ## original mesh file msh = meshio.read(mshfile) ## physical surface & volume data for key in msh.cell_data_dict["gmsh:physical"].keys(): if key == "triangle": triangle_data = msh.cell_data_dict["gmsh:physical"][key] elif key == "tetra": tetra_data = msh.cell_data_dict["gmsh:physical"][key] ## cell data tetra_cells = np.array([None]) triangle_cells = np.array([None]) for cell in msh.cells: if cell.type == "tetra": if tetra_cells.all() == None: tetra_cells = cell.data else: tetra_cells = np.concatenate((tetra_cells,cell.data)) elif cell.type == "triangle": if triangle_cells.all() == None: triangle_cells = cell.data else: triangle_cells = np.concatenate((triangle_cells,cell.data)) ## put them together tetra_mesh = meshio.Mesh(points=msh.points, cells={"tetra": tetra_cells}, cell_data={"name_to_read":[tetra_data]}) triangle_mesh =meshio.Mesh(points=msh.points, cells=[("triangle", triangle_cells)], cell_data={"name_to_read":[triangle_data]}) ## output meshio.write("mesh.xdmf", tetra_mesh) meshio.write("mf.xdmf", triangle_mesh)As a beginner, it took me a while until I realize I was not importing the >physical volume data in a code that is copied-and-pasted from the long thread >about meshio. The >often-quoted example
mesh = Mesh() with XDMFFile("mesh.xdmf") as infile: infile.read(mesh) mvc = MeshValueCollection("size_t", mesh, 2) with XDMFFile("mf.xdmf") as infile: infile.read(mvc, "name_to_read") mf = cpp.mesh.MeshFunctionSizet(mesh, mvc)is only looking for the
"name_to_read"inmf.xdmf, which returns >(according to theconvertMesh()above)triangle_data. This is, of >course, only returning the physical surface data rather than the physical volume >data which I want. Although I am not 100% sure about the treatment ofmvc, the function below >is what I am using to import the mesh (and it works for me):def importMesh(): ## Import mesh mesh = fn.Mesh() with fn.XDMFFile("mesh.xdmf") as infile: infile.read(mesh) ## Import material info (physical volume) mvc = fn.MeshValueCollection("size_t", mesh, 2) with fn.XDMFFile("mesh.xdmf") as infile: infile.read(mvc, "name_to_read") materials = fn.cpp.mesh.MeshFunctionSizet(mesh, mvc) ## Import boundary info (physical surface) # overwriting mvc because it's not really being used anywhere else mvc = fn.MeshValueCollection("size_t", mesh, 2) with fn.XDMFFile("mf.xdmf") as infile: infile.read(mvc, "name_to_read") boundaries = fn.cpp.mesh.MeshFunctionSizet(mesh, mvc) return mesh,materials,boundariesThis is what I wanted to do. The goal was to assign the density to each material >(
$\rho$ : 'rho') that are marked individually with physical volume id. This >density informationrhois used when evaluating the PDE. To do this, the >following class is used:class Rho(fn.UserExpression): def __init__(self,materials,volume_list,rho_list,**kwargs): super().__init__(**kwargs) self.materials = materials self.volume_list = volume_list self.rho_list = rho_list def value_shape(self): return () def eval_cell(self,values,x,cell): values[0] = 0 for i in range(len(self.volume_list)): if self.materials[cell.index] == self.volume_list[i]: values[0] = self.rho_list[i]Using above functions & a class, my code looks like this:
import fenics as fn import numpy as np import meshio # mesh prep convertMesh('meshfilename.msh') mesh,materials,boundaries = importMesh() # function space dx = fn.Measure('dx', domain=mesh, subdomain_data=mvc) V = fn.FunctionSpace(mesh, 'CG', 1) # boundary condition # (applying a pre-defined boundary value outer_value to physical surface #5) outer_boundary = fn.DirichletBC(V, outer_value, boundaries, 5) bcs = outer_boundary # material assignment # (applying three different values to physical volume #3,4, and 6) volume_list = [3,4,6] # list of physical volume ID's rho_list = [rho_vacuum,rho_inner,rho_outer] # list of pre-defined values rho = Rho(materials,volume_list,rho_list,degree=0) # solve (just an example: not really relevant) # solving a PDE for a scalar field phi (nonlinear Poisson) # pre-defined constants: initial_guess, Lambda, M, scaling phi = fn.project(initial_guess,V) v = fn.TestFunction(V) f = Lambda**5/phi**2 g = fn.Constant(0) a = (fn.inner(fn.grad(phi)*scaling**2,fn.grad(v))+ rho*v/M)/scaling*fn.dx L = f*v/scaling*fn.dx + g*v*fn.ds F = a-L fn.solve(F==0, phi, bcs)I hope this helps someone who is stuck at the same place as I was.