This project aims to build data synthesize systems to contaminate the clean DNA strands, to simulate the changes of DNA brought by the write and read operation of DNA-based storage systems. Specifically, three different methods are demonstrated:
- a naïve rule-based method
- A multi-layer perceptron network
- A sequence-to-sequence recurrent neural network
To test the quality of synthesized data, double-sided Bitwise Majority Alignment (BMA) algorithm is run on both generated data and real noisy data from DNA reads. The more similar the BMA algorithm behaves, the higher the generated data quality is.
In addition to the training and evaluation code, we also demonstrate some promising results: using the generated noisy strands from our seq2seq network, the given trace reconstruction algorithm behaves very similarly as when giving the real noisy data as input.
Trace reconstruction result comparison of real data and generated data by seq2seq model:
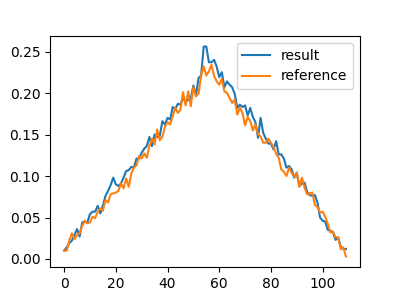
Other numeric metric:
Result on real data:
Average reconstruction error rate per position: 0.1181,
Number of perfectly reconstructed strands: 332,
Result on synthesized data:
Average reconstruction error rate per position: 0.1238,
Number of perfectly reconstructed strands: 338,
Average of positional absolute error rate difference: 0.0080
The structure of this repository is as follows:
.
├── data Directory for data
├── train.json Training split
├── valid.json Validation split
└── test.json Test split
├── dnacodec Implementation of BMA trace reconstruction
├── hparams Training configurations
├── other
├── mlp.yaml Config for mlp network
└── s2s_rnn.yaml Config for seq2seq network
├── models Implementation of network structure
├── results/ms_nano Results obtained by author
├── other
├── MLP
├── recon_ref
├── rule_based
└── Seq2seqRNN
├── checkpoint.pth Trained model
├── log.txt Training log
├── net_params.txt Printed network structure
├── recon_compare.json Behavior comparison of BMA
├── recon_compare.png Behavior comparison visualization
└── synthesized.json Synthesized strands
├── dataset.py Dataset class for network training
├── env.yaml Conda environment config
├── evaluate_recon.py Evaluate trace reconstruction results
├── format_data.py Format raw data into structured format
├── Readme.md
├── recon.py Compare trace reconstruction behavior
├── rule_based_method.py A rule_based system
├── train_s2s.py A seq2seq network-based system
├── train.py A multi-layer perceptron system
└── utils.py Tokenizer, early stopping, etc.
-
If the project folder doesn't have directory
data/Microsoft_NanoporeDownload the dataset from here. After downloading, put all contents of the downloaed folder to
data/Microsoft_Nanopore/raw -
Create a conda environment for this project
## Linux with CUDA conda env create -n [env_name] -f env.yaml ## OSX pip install -r requirements.txt pip install torch==2.0.0 torchvision==0.15.1 torchaudio==2.0.1 -
If the project folder doesn't have directory
data/Microsoft_Nanopore/train.jsonConvert the raw data into structured format by
python format_data.py -
There are three methods implemented for the simulation. To build the corresponding synthesis system, run one of the three commands below. Note: you may need to change the output directory in mlp.yaml or s2s_rnn.yaml before training a new model yourself, otherwise the existing results might be overwrote.
# Build and evaluate a rule-based system python rule_based_method.py # Train, inference with, and evaluate a multi-layer perceptron network python train.py hparams/mlp.yaml # Train, inference with, and evaluate a sequence-to-sequence network python train_s2s.py hparams/s2s_rnn.yaml
Please follow the two examples in infer.py file.
# In infer.py file
# Txt input, txt output
eg_txt()
# Json input, json output
eg_json()
MIT License, inherited from Microsoft_Nanopore.
-
Splitted dataset from Microsoft_Nanopore
-
The sequence-to-sequence network structure is adapted from [2] Bahdanau, Dzmitry, Kyung Hyun Cho, and Yoshua Bengio. "Neural machine translation by jointly learning to align and translate. " 3rd International Conference on Learning Representations, ICLR 2015. 2015.
-
Implementation of AttentionalRnnDecoder is adapted from SpeechBrain Toolkit: https://speechbrain.readthedocs.io/en/latest/API/speechbrain.nnet.RNN.html#speechbrain.nnet.RNN.AttentionalRNNDecoder