Image from the University of Cambridge
23andme once offered me a free DNA and ancestry test kit if I participated in one of their clinical studies. In exchange for a cheek swab and baring my guts and soul in a score of questionnaires, I got my genome sequenced and gained access to myriad reports on where my ancestors were likely from, whom else on the site I might be related to, and what health conditions and traits I probably have inherited.
Seriously?23andme already provides an overwhelming amount of consumer-ready infographics and tools, but I knew I could do more with the data. The intrepid may download their raw genetic data if they dare, so of course I poured it into pandas to see what I could make of it.
%matplotlib inline
import seaborn as sns
sns.set_style('darkgrid')
sns.color_palette('Spectral')
import matplotlib.pyplot as pltimport numpy as np
import requests
import pandas as pd
import refrom selenium import webdriver
from selenium.webdriver.support.ui import WebDriverWaitLooking at the .txt file, I could see that I was missing some genotype values, denoted with '--'.
Most of the chromosomes are ints, but three are X, Y, and MT (for 'mitochondrial'). I needed to specify the data type properly so that pandas wouldn't throw an error when it found mixed data in the input.
The other columns were fairly straightforward. I also wanted pandas to ignore the prefatory comments at the beginning of the file that consisted of lines beginning with an octothorpe.
The arguments I needed to pass, therefore, were:
- separator (tab-delimited)
- dtype (as a dict)
- na_values ('--') (n.b.: I decided against this in the end to avoid dealing with more NaNs)
- comment ('#')
data = pd.read_csv('genome.txt', sep='\t', dtype={'rsid':'str', 'chromosome':'object', 'position':'int', 'genotype':'str'}, comment='#')
print(data) rsid chromosome position genotype
0 rs548049170 1 69869 TT
1 rs13328684 1 74792 --
2 rs9283150 1 565508 AA
3 i713426 1 726912 --
4 rs116587930 1 727841 GG
5 rs3131972 1 752721 AG
6 rs12184325 1 754105 CC
... ... ... ... ...
[638531 rows x 4 columns]
A quick note on the column names:
-
rsid stands for Reference SNP cluster ID. It identifies unique SNPs.
-
SNPs are Single Nucleotide Polymorphisms ('snips'), locations in the genome that vary between individuals. They can influence disease risk and drug effects, tell you about your ancestry, and predict aspects of how you look and act.
-
All humans have almost the same sequence of 3 billion DNA bases (A,C,G, or T) distributed between their 23 pairs of chromosomes. But at certain locations, some differences exist that researchers have declared meaningful, for medical or other reasons (like genealogy).
I started to navigate my new DataFrame with basic exploratory data analysis and data cleaning.
# Read the data into a pandas DataFrame and do some EDA
df = pd.DataFrame(data)df.head(25).dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| rsid | chromosome | position | genotype | |
|---|---|---|---|---|
| 0 | rs548049170 | 1 | 69869 | TT |
| 1 | rs13328684 | 1 | 74792 | -- |
| 2 | rs9283150 | 1 | 565508 | AA |
| 3 | i713426 | 1 | 726912 | -- |
| 4 | rs116587930 | 1 | 727841 | GG |
| 5 | rs3131972 | 1 | 752721 | AG |
| 6 | rs12184325 | 1 | 754105 | CC |
| 7 | rs12567639 | 1 | 756268 | AA |
| 8 | rs114525117 | 1 | 759036 | GG |
| 9 | rs12124819 | 1 | 776546 | AA |
| 10 | rs12127425 | 1 | 794332 | GG |
| 11 | rs79373928 | 1 | 801536 | TT |
| 12 | rs72888853 | 1 | 815421 | -- |
| 13 | rs7538305 | 1 | 824398 | AA |
| 14 | rs28444699 | 1 | 830181 | AA |
| 15 | i713449 | 1 | 830731 | -- |
| 16 | rs116452738 | 1 | 834830 | GG |
| 17 | rs72631887 | 1 | 835092 | TT |
| 18 | rs28678693 | 1 | 838665 | TT |
| 19 | rs4970382 | 1 | 840753 | CT |
| 20 | rs4475691 | 1 | 846808 | CC |
| 21 | rs72631889 | 1 | 851390 | GG |
| 22 | rs7537756 | 1 | 854250 | AA |
| 23 | rs13302982 | 1 | 861808 | GG |
| 24 | rs376747791 | 1 | 863130 | AA |
df.isna().any() rsid False
chromosome False
position False
genotype False
dtype: bool
df.nunique() rsid 638531
chromosome 25
position 634934
genotype 20
dtype: int64
# How many chromosomes am I missing by not
# having a Y chromosome?
Y_chromosome = df[df.chromosome == 'Y']len(Y_chromosome)3733
I converted the letter chromosomes to numbers, cast them to ints, and created a dictionary to translate them back later so that I could better manipulate the data.
df['chromosome'].unique()array(['1', '2', '3', '4', '5', '6', '7', '8',
'9', '10', '11', '12','13', '14', '15',
'16', '17', '18', '19', '20', '21', '22',
'X','MT'], dtype=object)
df['chromosome'] = df['chromosome'].apply(lambda x:
re.sub(r'X', r'23', x))
df['chromosome'] = df['chromosome'].apply(lambda x:
re.sub(r'MT', r'24', x))df['chromosome'] = df['chromosome'].apply(lambda x:
int(x))chromosome_dict = {1:'1', 2:'2', 3:'3', 4:'4', 5:'5',
6:'6', 7:'7', 8:'8', 9:'9', 10:'10',
11:'11', 12:'12', 13:'13', 14:'14',
15:'15', 16:'16', 17:'17', 18:'18',
19:'19', 20:'20', 21:'21', 22:'22',
23:'X', 24:'MT'}print(chromosome_dict)
display(df.info()){1: '1', 2: '2', 3: '3', 4: '4', 5: '5', 6: '6', 7: '7',
8: '8', 9: '9', 10: '10', 11: '11', 12: '12', 13: '13',
14: '14', 15: '15', 16: '16', 17: '17', 18: '18',
19: '19', 20: '20', 21: '21', 22: '22', 23: 'X',
24: 'MT'}
<class 'pandas.core.frame.DataFrame'>
Int64Index: 634798 entries, 0 to 638530
Data columns (total 4 columns):
rsid 634798 non-null object
chromosome 634798 non-null int64
position 634798 non-null int64
genotype 634798 non-null object
dtypes: int64(2), object(2)
memory usage: 24.2+ MB
None
There were 16,005 genotypes that I simply lacked:
genotype_na = df[df.genotype == '--']
len(genotype_na)16005
df[df.chromosome == 1].info()<class 'pandas.core.frame.DataFrame'>
Int64Index: 49514 entries, 0 to 49513
Data columns (total 4 columns):
rsid 49514 non-null object
chromosome 49514 non-null int64
position 49514 non-null int64
genotype 49514 non-null object
dtypes: int64(2), object(2)
memory usage: 1.9+ MB
# Remove that pesky whitespace from the column name
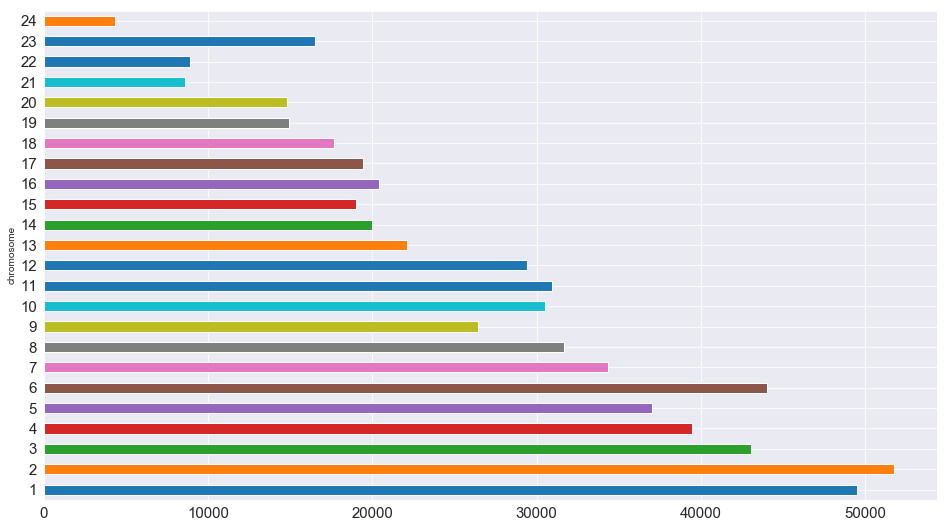
df.rename({' rsid': 'rsid'}, axis='columns', inplace=True)How many SNPs are there per chromosome?
# We can do this manually with a for loop . . .
x = []
y = []
for k in chromosome_dict:
x.append(k)
y.append(len(df[df.chromosome == k]))
rsid_per_chromosome = dict(zip(x,y)) rsid_per_chromosome{1: 49514,
2: 51775,
3: 43024,
4: 39474,
5: 37032,
6: 44023,
7: 34357,
8: 31683,
9: 26446,
10: 30525,
11: 30942,
12: 29432,
13: 22080,
14: 19961,
15: 19006,
16: 20397,
17: 19401,
18: 17675,
19: 14917,
20: 14781,
21: 8607,
22: 8915,
23: 16530,
24: 4301}
# . . . but pandas makes it a lot easier!
rsid_per_chromosome_series = df.groupby('chromosome')['rsid'].count()
rsid_per_chromosome_series.columns = ['chromosome', 'count']rsid_per_chromosome_series.plot.barh(figsize=(16,9), fontsize=15)
plt.show()To acquire more information about my DNA, I pulled files from SNPedia, a wiki investigating human genetics that gathers extensive data and cites to peer-reviewed scientific publications. SNPedia catalogues common, reproducible SNPs (or ones found in meta-analyses or studies of at least 500 patients), or those with other historic or medical significance.
The columns are:
- Unnamed: 0 (actually the SNP name)
- Magnitude (a subjective measure of interest)
- Repute (a subjective measure of whether the genotype is "good" or "bad" to have based on research, and blank for things like ancestry and eye color)
- Summary (a narrative description)
snp_df = pd.read_csv('result.csv')
snp_df.head().dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| Unnamed: 0 | Magnitude | Repute | Summary | |
|---|---|---|---|---|
| 0 | Rs661(A;A) | 9.0 | Bad | early onset Alzheimer's disease |
| 1 | Rs6647(T;T) | 0.0 | Good | Normal; two copies of Pi-M1V allele |
| 2 | Rs6647(C;C) | 0.0 | Good | Normal; two copies of Pi-M1A allele |
| 3 | Rs1303(T;T) | 0.0 | Good | common in clinvar |
| 4 | Rs28929471(G;G) | 0.0 | Good | common in complete genomics |
To align with my original DataFrame, I created a genotype column and used regex to separate out the genotype, which was stitched onto the end of the SNP name.
snp_df['genotype'] = snp_df['Unnamed: 0'].apply(lambda x:
re.sub(r'.*([AGCT]);([AGCT])\)', r'\1\2', x))snp_df.head().dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| Unnamed: 0 | Magnitude | Repute | Summary | genotype | |
|---|---|---|---|---|---|
| 0 | Rs661(A;A) | 9.0 | Bad | early onset Alzheimer's disease | AA |
| 1 | Rs6647(T;T) | 0.0 | Good | Normal; two copies of Pi-M1V allele | TT |
| 2 | Rs6647(C;C) | 0.0 | Good | Normal; two copies of Pi-M1A allele | CC |
| 3 | Rs1303(T;T) | 0.0 | Good | common in clinvar | TT |
| 4 | Rs28929471(G;G) | 0.0 | Good | common in complete genomics | GG |
For consistency's sake, I renamed the columns to match my original DataFrame and made sure the rsids were all lower-case.
new_cols = ['rsid', 'magnitude', 'repute',
'summary', 'genotype']
snp_df.columns = new_colsI used regex to clean up the rsid a little more, too (because I will take any excuse to use more regex).
snp_df['rsid'] = snp_df['rsid'].map(lambda x : x.lower())
snp_df['rsid'] = snp_df['rsid'].map(lambda x :
re.sub(r'([a-z]{1,}[\d]+)\([agct];[agct]\)',
r'\1', x))snp_df.head().dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| rsid | magnitude | repute | summary | genotype | |
|---|---|---|---|---|---|
| 0 | rs661 | 9.0 | Bad | early onset Alzheimer's disease | AA |
| 1 | rs6647 | 0.0 | Good | Normal; two copies of Pi-M1V allele | TT |
| 2 | rs6647 | 0.0 | Good | Normal; two copies of Pi-M1A allele | CC |
| 3 | rs1303 | 0.0 | Good | common in clinvar | TT |
| 4 | rs28929471 | 0.0 | Good | common in complete genomics | GG |
snp_df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 100 entries, 0 to 99
Data columns (total 5 columns):
rsid 100 non-null object
magnitude 100 non-null float64
repute 91 non-null object
summary 96 non-null object
genotype 100 non-null object
dtypes: float64(1), object(4)
memory usage: 4.0+ KB
I overwrote the null reputes and summaries.
null_repute = snp_df[snp_df['repute'].isnull()]
null_summaries = snp_df[snp_df['summary'].isnull()]
null_repute_and_summaries = pd.concat([null_repute,null_summaries]).drop_duplicates().reset_index(drop=True)
display(null_repute_and_summaries).dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| rsid | magnitude | repute | summary | genotype | |
|---|---|---|---|---|---|
| 0 | rs28931569 | 4.0 | NaN | high risk of emphysema | CC |
| 1 | rs28929473 | 0.0 | NaN | NaN | TT |
| 2 | rs28931572 | 0.0 | NaN | NaN | AA |
| 3 | rs1801252 | 3.0 | NaN | NaN | GG |
| 4 | rs8192466 | 4.0 | NaN | uncertain | TT |
| 5 | rs4986852 | 2.0 | NaN | predisposition to breast cancer? | AA |
| 6 | rs1800709 | 2.0 | NaN | predisposition to breast cancer? | TT |
| 7 | rs28931592 | 0.0 | NaN | NaN | TT |
| 8 | rs4986893 | 2.1 | NaN | poor metabolizer of several commonly prescribe... | AA |
snp_df['repute'].fillna(value='Neutral', inplace=True)
snp_df['summary'].fillna(value='None', inplace=True)# No no NaNette
snp_df.isna().any()rsid False
magnitude False
repute False
summary False
genotype False
dtype: bool
Appropriately enough, I did an inner join of the SNPedia DataFrame on my DNA to see what data, if any, it had on my particular genotypes.
new_df = snp_df.merge(df, how='inner', on=['rsid', 'genotype'], suffixes=('_SNPedia', '_myDNA'))new_df.head(20).dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| rsid | magnitude | repute | summary | genotype | chromosome | position | |
|---|---|---|---|---|---|---|---|
| 0 | rs1303 | 0.0 | Good | common in clinvar | TT | 14 | 94844843 |
| 1 | rs17580 | 2.5 | Bad | a slightly reduced functionality form of Alpha... | TT | 14 | 94847262 |
| 2 | rs28931580 | 0.0 | Good | common in clinvar | AA | 12 | 50344783 |
| 3 | rs1042714 | 0.0 | Good | normal | CC | 5 | 148206473 |
| 4 | rs1800888 | 0.0 | Good | normal | CC | 5 | 148206885 |
| 5 | rs2303790 | 0.0 | Good | common in clinvar | AA | 16 | 57017292 |
| 6 | rs5882 | 2.0 | Bad | Faster aging. Increased risk for Dementia. Les... | AA | 16 | 57016092 |
| 7 | rs2230199 | 2.0 | Bad | 2.5x+ risk of ARMD | GG | 19 | 6718387 |
| 8 | rs28931608 | 0.0 | Good | common in clinvar | GG | 7 | 75614497 |
| 9 | rs4986893 | 0.0 | Good | normal | GG | 10 | 96540410 |
| 10 | rs28399504 | 0.0 | Good | normal | AA | 10 | 96522463 |
| 11 | rs2234922 | 0.0 | Good | common in clinvar | AA | 1 | 226026406 |
| 12 | rs28931614 | 0.0 | Good | common in clinvar | GG | 4 | 1806119 |
| 13 | rs28933068 | 0.0 | Good | common in clinvar | CC | 4 | 1807371 |
# Create a DataFrame for some subsets of genes
good_genes = new_df[new_df.repute == 'Good']
bad_genes = new_df[new_df.repute == 'Bad']
interesting_genes = new_df[new_df.magnitude > 4] # 4 is the threshold for "worth your time" given by SNPediaI have plenty of "good" genotypes, but none with a nonzero magnitude.
good_genes.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| rsid | magnitude | repute | summary | genotype | chromosome | position | |
|---|---|---|---|---|---|---|---|
| 0 | rs1303 | 0.0 | Good | common in clinvar | TT | 14 | 94844843 |
| 2 | rs28931580 | 0.0 | Good | common in clinvar | AA | 12 | 50344783 |
| 3 | rs1042714 | 0.0 | Good | normal | CC | 5 | 148206473 |
| 4 | rs1800888 | 0.0 | Good | normal | CC | 5 | 148206885 |
| 5 | rs2303790 | 0.0 | Good | common in clinvar | AA | 16 | 57017292 |
| 8 | rs28931608 | 0.0 | Good | common in clinvar | GG | 7 | 75614497 |
| 9 | rs4986893 | 0.0 | Good | normal | GG | 10 | 96540410 |
| 10 | rs28399504 | 0.0 | Good | normal | AA | 10 | 96522463 |
| 11 | rs2234922 | 0.0 | Good | common in clinvar | AA | 1 | 226026406 |
| 12 | rs28931614 | 0.0 | Good | common in clinvar | GG | 4 | 1806119 |
| 13 | rs28933068 | 0.0 | Good | common in clinvar | CC | 4 | 1807371 |
I have three "bad" genotypes with a nonzero magnitude.
bad_genes.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| rsid | magnitude | repute | summary | genotype | chromosome | position | |
|---|---|---|---|---|---|---|---|
| 1 | rs17580 | 2.5 | Bad | a slightly reduced functionality form of Alpha... | TT | 14 | 94847262 |
| 6 | rs5882 | 2.0 | Bad | Faster aging. Increased risk for Dementia. Les... | AA | 16 | 57016092 |
| 7 | rs2230199 | 2.0 | Bad | 2.5x+ risk of ARMD | GG | 19 | 6718387 |
Sadly, I had no "interesting" genotypes above the threshold of 4, although hearteningly I did possess some slightly interesting bad ones.
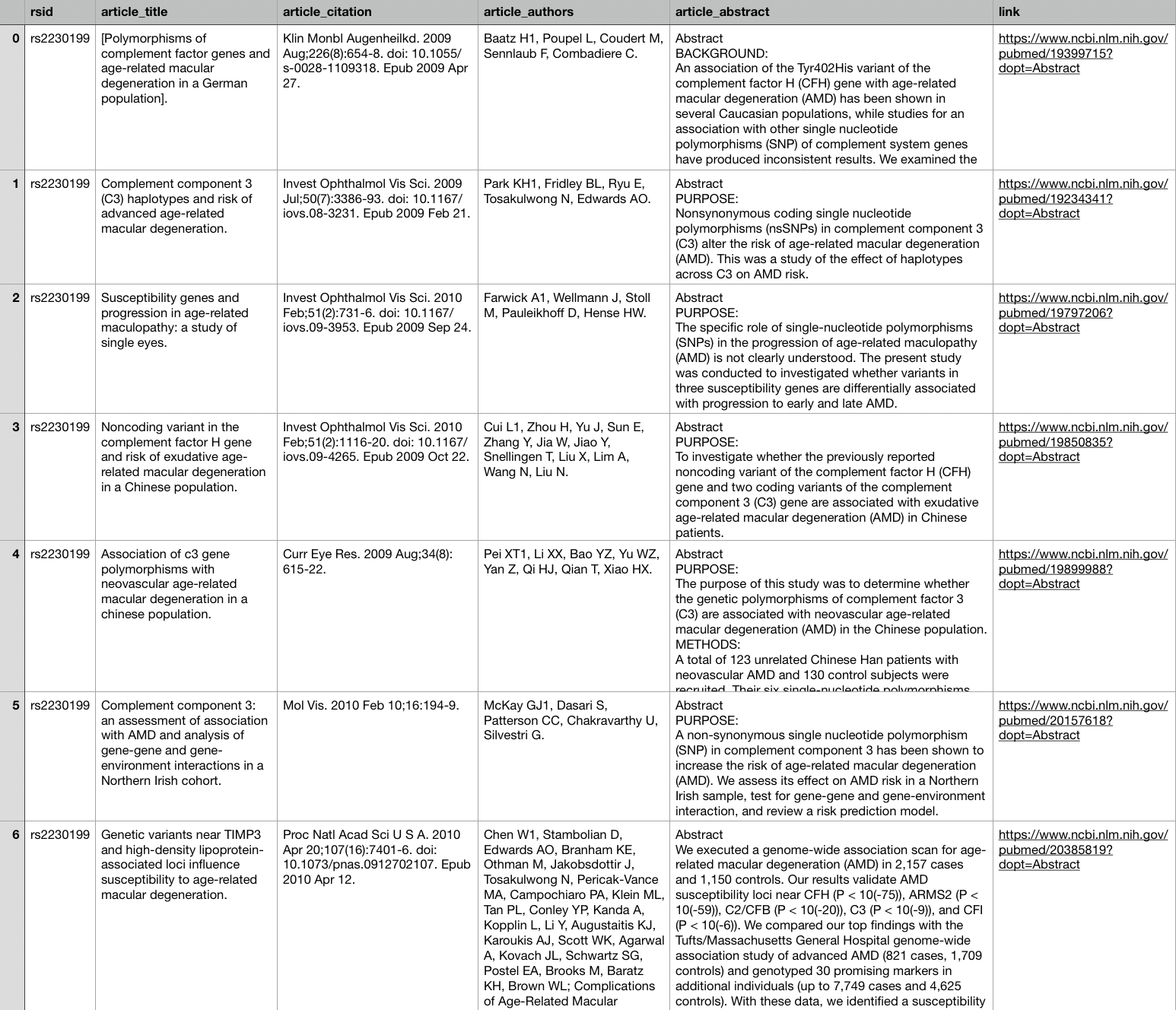
I decided I might like to read up on my bad genetics, so I used Selenium to scrape the abstracts of some scientific papers from PubMed.
# Get the base URL from SNPedia
base_url = 'https://www.snpedia.com/index.php/'# Create URLs for each gene that I want to study
gene_urls = [base_url + rsid for rsid in bad_genes['rsid']]# Initialize Selenium
browser = webdriver.Chrome()import time# Write a function to visit the SNPedia URLs, click through to PubMed,
# and retrieve the info on the articles for each gene
def scrape_abstracts(urls):
rsid_list = []
all_article_title = []
all_article_citation = []
all_article_authors = []
all_article_abstract = []
all_article_links = []
for url in urls:
link_urls = []
browser.get(url) #load url
rsid = browser.find_element_by_css_selector('.firstHeading').text
links_elements = browser.find_elements_by_partial_link_text('PMID')
# get the URLs to the PubMed pages
for link in links_elements:
link_urls.append(link.get_attribute('href'))
# follow each link element to PubMed
for element in link_urls:
browser.get(element)
time.sleep(2)
article_title = browser.find_element_by_xpath("//div[@class='cit']/../h1").text
article_citation = browser.find_element_by_class_name('cit').text
article_authors = browser.find_element_by_class_name('auths').text
article_abstract = browser.find_element_by_class_name('abstr').text
rsid_list.append(rsid)
all_article_title.append(article_title)
all_article_citation.append(article_citation)
all_article_authors.append(article_authors)
all_article_abstract.append(article_abstract)
all_article_links.append(element)
# store the information
df = pd.DataFrame()
df['rsid'] = rsid_list
df['article_title'] = all_article_title
df['article_citation'] = all_article_citation
df['article_authors'] = all_article_authors
df['article_abstract'] = all_article_abstract
df['link'] = all_article_links
df = df.drop_duplicates()
df.index = range(len(df.index))
return dfabstracts_df = scrape_abstracts(gene_urls)For later hypochondriacal perusal, I exported my findings, complete with abstracts and hyperlinks, to a CSV file using the pandas DataFrame.to_csv method.
# DataFrame to CSV
export_csv = abstracts_df.to_csv(r'/Users/lorajohns/Documents/Python/DNA/DNA_articles.csv')Now I have a handy CSV file, nicely formatted, with citations to scientific articles analyzing and describing my probably un-problematic, but probationally proditory genotypes. Python provides prodigious tools to engage in literal introspection that the sawbones of old could never have imagined.