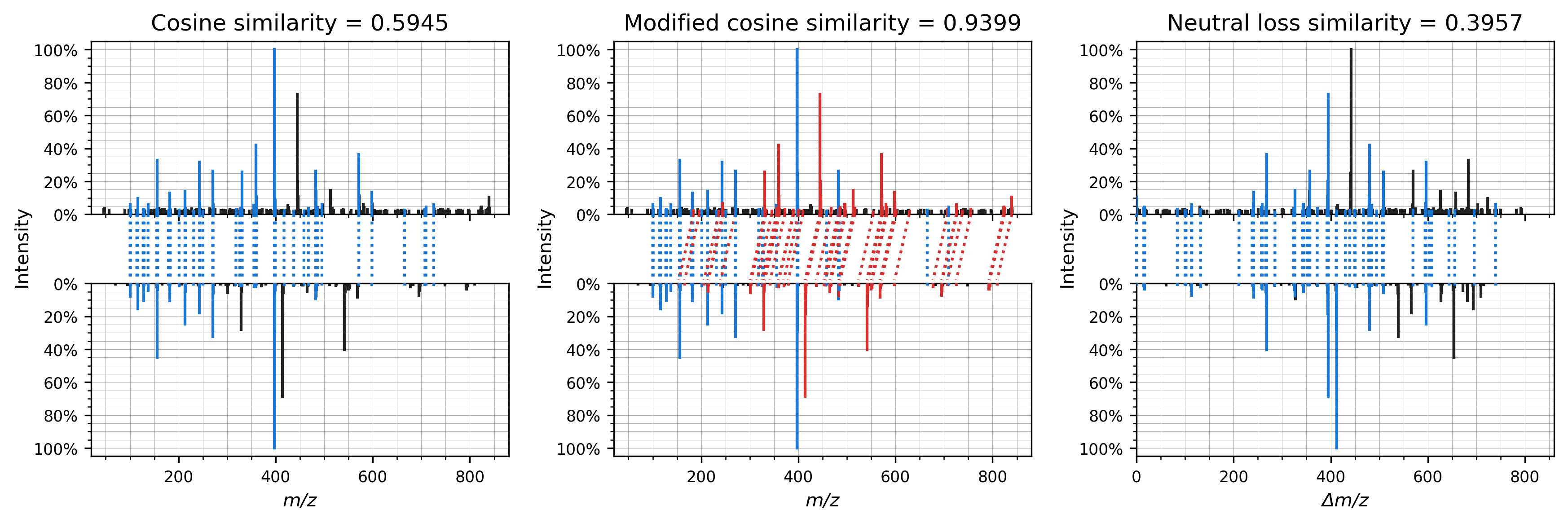
This repository contains code to compare several spectrum similarity measures for the discovery of structurally related molecules.
Similarity measures that are currently implemented are:
- Cosine similarity
Stein, S. E. & Scott, D. R. Optimization and testing of mass spectral library search algorithms for compound identification. Journal of the American Society for Mass Spectrometry 5, 859–866 (1994). doi:10.1016/1044-0305(94)87009-8 - Modified cosine similarity
Watrous, J. et al. Mass spectral molecular networking of living microbial colonies. Proceedings of the National Academy of Sciences 109, E1743–E1752 (2012). doi:10.1073/pnas.1203689109 - Neutral loss similarity
Aisporna, A. et al. Neutral loss mass spectral data enhances molecular similarity analysis in METLIN. Journal of the American Society for Mass Spectrometry 33, 530–534 (2022). doi:10.1021/jasms.1c00343
All code is available as open-source under the BSD license.
If you use this software in your work, please cite the following publication:
- Bittremieux, W. et al. Comparison of cosine, modified cosine, and neutral loss based spectral alignment for discovery of structurally related molecules. bioRxiv (2022) doi:10.1101/2022.06.01.494370.
Create a conda environment and install all required dependencies using the provided environment.yml file:
conda env create -f https://raw.githubusercontent.com/bittremieux/cosine_neutral_loss/master/environment.yml
Code example to create mirror plots using different similarity measures:
import spectrum_utils.spectrum as sus
import plot
# Apratoxin A.
usi1 = "mzspec:GNPS:GNPS-LIBRARY:accession:CCMSLIB00000424840"
# Apratoxin A analog.
usi2 = "mzspec:MSV000086109:BD5_dil2x_BD5_01_57213:scan:760"
spectrum1 = sus.MsmsSpectrum.from_usi(usi1)
spectrum1.precursor_charge = 1
spectrum1 = spectrum1.set_mz_range(0, spectrum1.precursor_mz)
spectrum1 = spectrum1.remove_precursor_peak(0.1, "Da")
spectrum2 = sus.MsmsSpectrum.from_usi(usi2)
spectrum2.precursor_charge = 1
spectrum2 = spectrum2.set_mz_range(0, spectrum2.precursor_mz)
spectrum2 = spectrum2.remove_precursor_peak(0.1, "Da")
for score, filename in [
(None, "spectra.png"),
("cosine", "cosine.png"),
("modified_cosine", "modified_cosine.png"),
("neutral_loss", "neutral_loss.png"),
]:
plot.plot_mirror(spectrum1, spectrum2, score, filename)
For more advanced functionality, see the source code.
For more information you can visit the official code website or send an email to wbittremieux@health.ucsd.edu.