This is an implementation of the expectation maximization (EM) algorithm for maximum-likelihood estimation of Gaussian mixture model parameter estimation. It supports data of arbitrary dimensions, and avoids numerical errors in the presence of thin subspaces or degenerate data by clamping the eigenvalues of the covariance matrices via a user-defined threshold. It depends on the Eigen linear algebra library.
To use this code, just include the source files in gmm/ in your project, and #include "GaussianMixture.h".
The below snippet initializes a Gaussian mixture model with 3 components in 2 dimensions, with a minimum component variance of 0.01, then optimizes the parameters for 50 random observations and reports the likelihood of a new observation given the learned model.
GaussianMixture gmm(3, 2, 0.01);
Eigen::MatrixXd data = Eigen::MatrixXd::Random(50, 2);
int iters = gmm.learn(data);
std::cout << "took " << iters << " iterations" << std::endl;
std::cout << "-------- parameters --------" << std::endl << gmm << std::endl;
Eigen::RowVector2d point(0, 0);
std::cout << "log likelihood at point " << point << ": " << gmm.logp_data(point) << std::endl;The below example shows how to manually set the parameters of the model and use it as a likelihood function:
GaussianMixture gmm;
std::vector<Eigen::MatrixXd> cov_default(1, Eigen::MatrixXd::Identity(3, 3));
Eigen::Matrix<double, 1, 1> pi(1);
Eigen::Matrix<double, 1, 2> mean(1, -1);
gmm.initialize(mean, cov_default, pi);
gmm.useCurrentModel();
Eigen::RowVector2d point(0, 0);
std::cout << "log likelihood at point " << point << ": " << gmm.logp_data(point) << std::endl;See `gmm/GaussianMixture.h" for more detailed documentation on all the functions.
To build the example tests, navigate to the root directory, and run
mkdir build
cd build
cmake .. -DCMAKE_BUILD_TYPE=Release
make
It is important to build in release mode, since Eigen is much slower in debug mode.
If you have OpenCV installed, you can enable rendering the results to images:
cmake .. -DCMAKE_BUILD_TYPE=Release -DUSE_OPENCV=on
make
Run the tests with
./GMMTests [num_comps]
where num_comps is the number of components to generate and attempt to recover the parameters for.
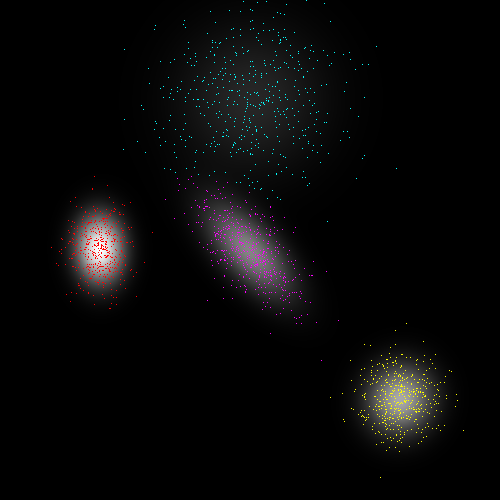
The test application generates 2D points in a plane according to a mixture of gaussians, then attempts to recover the mixture parameters from the observed samples. It also tests the numerical stability in the presence of thin subspaces and degenerate data. Below are the visualizations of typical test results (these can be obtained by enabling the USE_OPENCV flag in the cmake options). Colored points are training data, colored by maximum component likelihood, and the shading is the likelihood of each pixel given the model.