rgbif
rgbif gives you access to data from GBIF via their REST API. GBIF versions their API - we are currently using v1 of their API. You can no longer use their old API in this package - see ?rgbif-defunct.
Installation
install.packages("rgbif")Alternatively, install development version
install.packages("devtools")
devtools::install_github("ropensci/rgbif")library("rgbif")Note: Windows users have to first install Rtools to use devtools
Search for occurrence data
occ_search(scientificName = "Ursus americanus", limit = 50)
#> Records found [7218]
#> Records returned [50]
#> No. unique hierarchies [1]
#> No. media records [50]
#> Args [scientificName=Ursus americanus, limit=50, offset=0, fields=all]
#> First 10 rows of data
#>
#> name key decimalLatitude decimalLongitude
#> 1 Ursus americanus 1065590124 38.36662 -79.68283
#> 2 Ursus americanus 1065588899 35.73304 -82.42028
#> 3 Ursus americanus 1065611122 43.94883 -72.77432
#> 4 Ursus americanus 1098894889 23.69470 -99.14630
#> 5 Ursus americanus 1088908315 43.86464 -72.34617
#> 6 Ursus americanus 1088923534 36.93018 -78.25027
#> 7 Ursus americanus 1088932238 32.65219 -108.53674
#> 8 Ursus americanus 1088932273 32.65237 -108.53691
#> 9 Ursus americanus 1088964797 29.27042 -103.30058
#> 10 Ursus americanus 1088961422 49.72317 -96.03215
#> .. ... ... ... ...
#> Variables not shown: issues (chr), datasetKey (chr), publishingOrgKey
#> (chr), publishingCountry (chr), protocol (chr), lastCrawled (chr),
#> lastParsed (chr), extensions (chr), basisOfRecord (chr), taxonKey
#> (int), kingdomKey (int), phylumKey (int), classKey (int), orderKey
#> (int), familyKey (int), genusKey (int), speciesKey (int),
#> scientificName (chr), kingdom (chr), phylum (chr), order (chr),
#> family (chr), genus (chr), species (chr), genericName (chr),
#> specificEpithet (chr), taxonRank (chr), dateIdentified (chr), year
#> (int), month (int), day (int), eventDate (chr), modified (chr),
#> lastInterpreted (chr), references (chr), identifiers (chr), facts
#> (chr), relations (chr), geodeticDatum (chr), class (chr), countryCode
#> (chr), country (chr), verbatimEventDate (chr),
#> http...unknown.org.occurrenceDetails (chr), rights (chr),
#> rightsHolder (chr), occurrenceID (chr), taxonID (chr), collectionCode
#> (chr), gbifID (chr), occurrenceRemarks (chr), institutionCode (chr),
#> datasetName (chr), catalogNumber (chr), recordedBy (chr), eventTime
#> (chr), identifier (chr), identificationID (chr), verbatimLocality
#> (chr), infraspecificEpithet (chr), informationWithheld (chr)Or you can get the taxon key first with name_backbone(). Here, we select to only return the occurrence data.
key <- name_backbone(name='Helianthus annuus', kingdom='plants')$speciesKey
occ_search(taxonKey=key, limit=20)
#> Records found [20650]
#> Records returned [20]
#> No. unique hierarchies [1]
#> No. media records [18]
#> Args [taxonKey=3119195, limit=20, offset=0, fields=all]
#> First 10 rows of data
#>
#> name key decimalLatitude decimalLongitude
#> 1 Helianthus annuus 1095851641 0.00000 0.00000
#> 2 Helianthus annuus 1088900309 33.95239 -117.32011
#> 3 Helianthus annuus 1088933055 25.66564 -100.30348
#> 4 Helianthus annuus 1088909392 24.72449 -99.54020
#> 5 Helianthus annuus 1088937716 25.81691 -100.05940
#> 6 Helianthus annuus 1088944416 26.20518 -98.26725
#> 7 Helianthus annuus 1090389390 59.96150 17.71060
#> 8 Helianthus annuus 1092889365 32.71840 -114.75603
#> 9 Helianthus annuus 1092889645 1.27617 103.79136
#> 10 Helianthus annuus 1098903927 29.17958 -102.99551
#> .. ... ... ... ...
#> Variables not shown: issues (chr), datasetKey (chr), publishingOrgKey
#> (chr), publishingCountry (chr), protocol (chr), lastCrawled (chr),
#> lastParsed (chr), extensions (chr), basisOfRecord (chr), taxonKey
#> (int), kingdomKey (int), phylumKey (int), classKey (int), orderKey
#> (int), familyKey (int), genusKey (int), speciesKey (int),
#> scientificName (chr), kingdom (chr), phylum (chr), order (chr),
#> family (chr), genus (chr), species (chr), genericName (chr),
#> specificEpithet (chr), taxonRank (chr), dateIdentified (chr),
#> elevation (dbl), elevationAccuracy (dbl), stateProvince (chr), year
#> (int), month (int), day (int), eventDate (chr), lastInterpreted
#> (chr), identifiers (chr), facts (chr), relations (chr), geodeticDatum
#> (chr), class (chr), countryCode (chr), country (chr), recordNumber
#> (chr), rights (chr), municipality (chr), rightsHolder (chr),
#> ownerInstitutionCode (chr), type (chr), occurrenceID (chr),
#> collectionCode (chr), identifiedBy (chr), gbifID (chr),
#> occurrenceRemarks (chr), institutionCode (chr), datasetName (chr),
#> catalogNumber (chr), recordedBy (chr), locality (chr), language
#> (chr), identifier (chr), modified (chr), references (chr),
#> verbatimEventDate (chr), verbatimLocality (chr),
#> http...unknown.org.occurrenceDetails (chr), taxonID (chr), eventTime
#> (chr), identificationID (chr), coordinateAccuracy (dbl), depth (dbl),
#> depthAccuracy (dbl), county (chr), informationWithheld (chr)Search for many species
Get the keys first with name_backbone(), then pass to occ_search()
splist <- c('Accipiter erythronemius', 'Junco hyemalis', 'Aix sponsa')
keys <- sapply(splist, function(x) name_backbone(name=x)$speciesKey, USE.NAMES=FALSE)
occ_search(taxonKey=keys, limit=5, hasCoordinate=TRUE)
#> Occ. found [2480598 (20), 2492010 (1925675), 2498387 (589944)]
#> Occ. returned [2480598 (5), 2492010 (5), 2498387 (5)]
#> No. unique hierarchies [2480598 (1), 2492010 (1), 2498387 (1)]
#> No. media records [2480598 (1), 2492010 (5), 2498387 (5)]
#> Args [taxonKey=2480598,2492010,2498387, hasCoordinate=TRUE, limit=5,
#> offset=0, fields=all]
#> First 10 rows of data from 2480598
#>
#> name key decimalLatitude decimalLongitude
#> 1 Accipiter erythronemius 920169861 -20.55244 -56.64104
#> 2 Accipiter erythronemius 920184036 -20.76029 -56.71314
#> 3 Accipiter erythronemius 1001096527 -27.58000 -58.66000
#> 4 Accipiter erythronemius 1001096518 -27.92000 -59.14000
#> 5 Accipiter erythronemius 699417490 5.26667 -60.73333
#> Variables not shown: issues (chr), datasetKey (chr), publishingOrgKey
#> (chr), publishingCountry (chr), protocol (chr), lastCrawled (chr),
#> lastParsed (chr), extensions (chr), basisOfRecord (chr), taxonKey
#> (int), kingdomKey (int), phylumKey (int), classKey (int), orderKey
#> (int), familyKey (int), genusKey (int), speciesKey (int),
#> scientificName (chr), kingdom (chr), phylum (chr), order (chr),
#> family (chr), genus (chr), species (chr), genericName (chr),
#> specificEpithet (chr), taxonRank (chr), year (int), month (int), day
#> (int), eventDate (chr), lastInterpreted (chr), identifiers (chr),
#> facts (chr), relations (chr), geodeticDatum (chr), class (chr),
#> countryCode (chr), country (chr), gbifID (chr), institutionCode
#> (chr), catalogNumber (chr), recordedBy (chr), locality (chr),
#> collectionCode (chr), modified (chr), created (chr),
#> higherClassification (chr), associatedSequences (chr), occurrenceID
#> (chr), identifier (chr), taxonID (chr), sex (chr), elevation (dbl),
#> elevationAccuracy (dbl), recordNumber (chr), institutionID (chr),
#> rights (chr), higherGeography (chr), type (chr), georeferenceSources
#> (chr), otherCatalogNumbers (chr), bibliographicCitation (chr),
#> verbatimEventDate (chr), preparations (chr),
#> georeferenceVerificationStatus (chr), occurrenceRemarks (chr),
#> accessRights (chr), datasetName (chr), verbatimElevation (chr),
#> language (chr)Maps
Make a simple map of species occurrences.
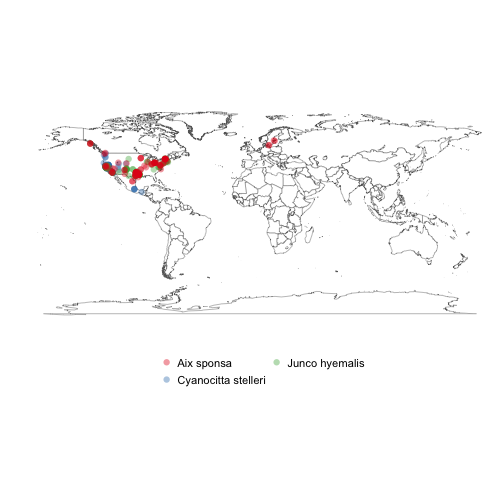
splist <- c('Cyanocitta stelleri', 'Junco hyemalis', 'Aix sponsa')
keys <- sapply(splist, function(x) name_backbone(name=x)$speciesKey, USE.NAMES=FALSE)
dat <- occ_search(taxonKey=keys, limit=100, return='data', hasCoordinate=TRUE)
library('plyr')
datdf <- ldply(dat)
gbifmap(datdf)Meta
- Please report any issues or bugs.
- License: MIT
- Get citation information for
rgbifin R doingcitation(package = 'rgbif')
This package is part of a richer suite called SPOCC Species Occurrence Data, along with several other packages, that provide access to occurrence records from multiple databases.