This repository provides benchmarked property prediction results on a curated dataset of 405 photoswitch molecules.
We recommend using a conda virtual environment.
conda create -n photoswitch python=3.7
conda install -c conda-forge rdkit
conda install seaborn xlrd matplotlib ipython pytest pytorch scikit-learn pandas
pip install git+https://github.com/GPflow/GPflow.git@develop#egg=gpflow
pip install dgl dgllife jupyter gpflux umap-learn
git clone https://github.com/hyperopt/hyperopt-sklearn.git
cd hyperopt-sklearn
pip install -e .
To reproduce the property prediction results, run a model prediction script using the task flag to specify the appropriate task (e_iso_pi, e_iso_n, z_iso_pi, z_iso_n) corresponding to different electronic transition wavelengths e.g.
python predict_with_GPR.py -task e_iso_pi | Metric | GPR-Tanimoto + Fragprints |
|---|---|
| RMSE | 20.9 nm |
| MAE | 13.3 nm |
| R2 | 0.90 |
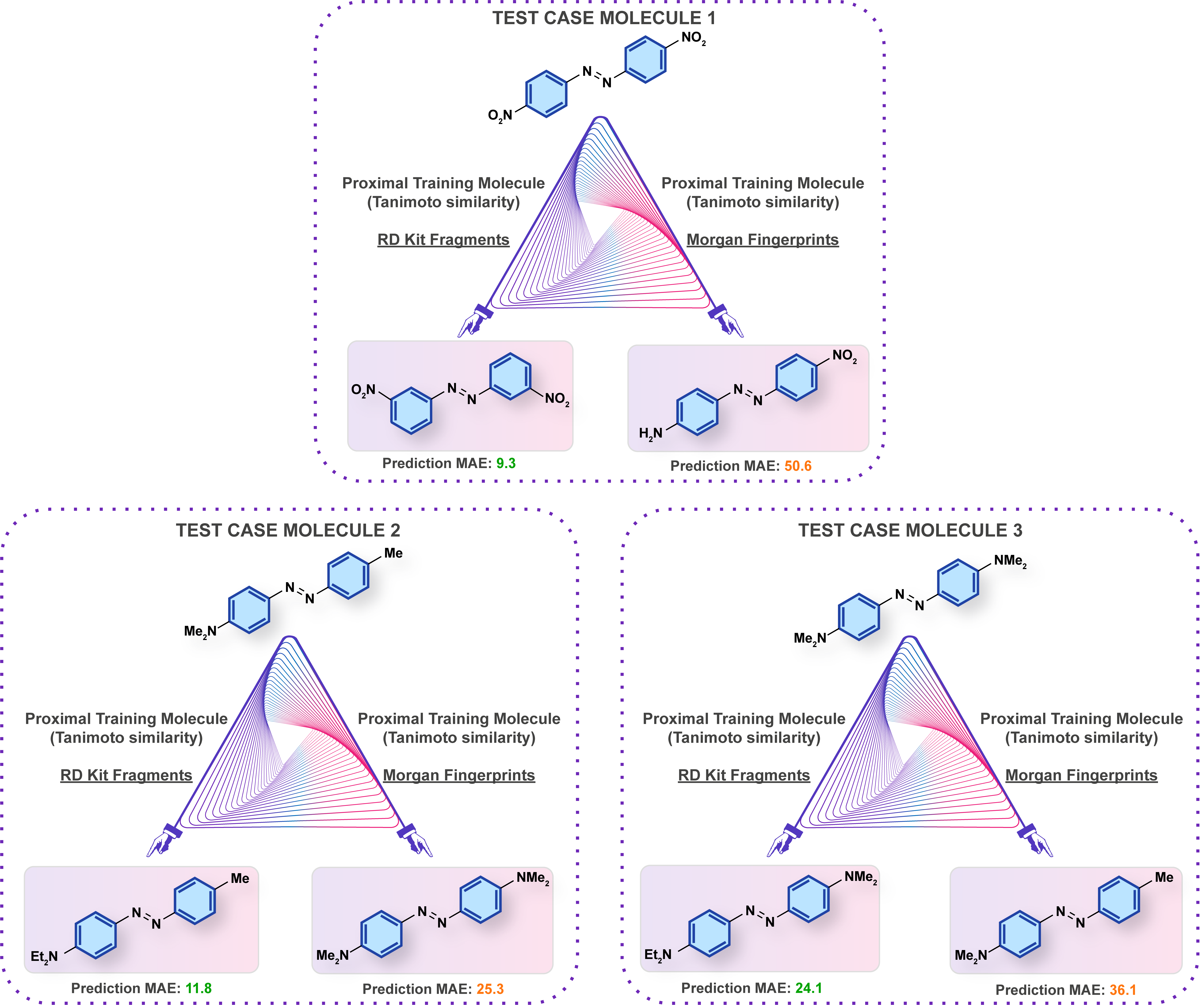
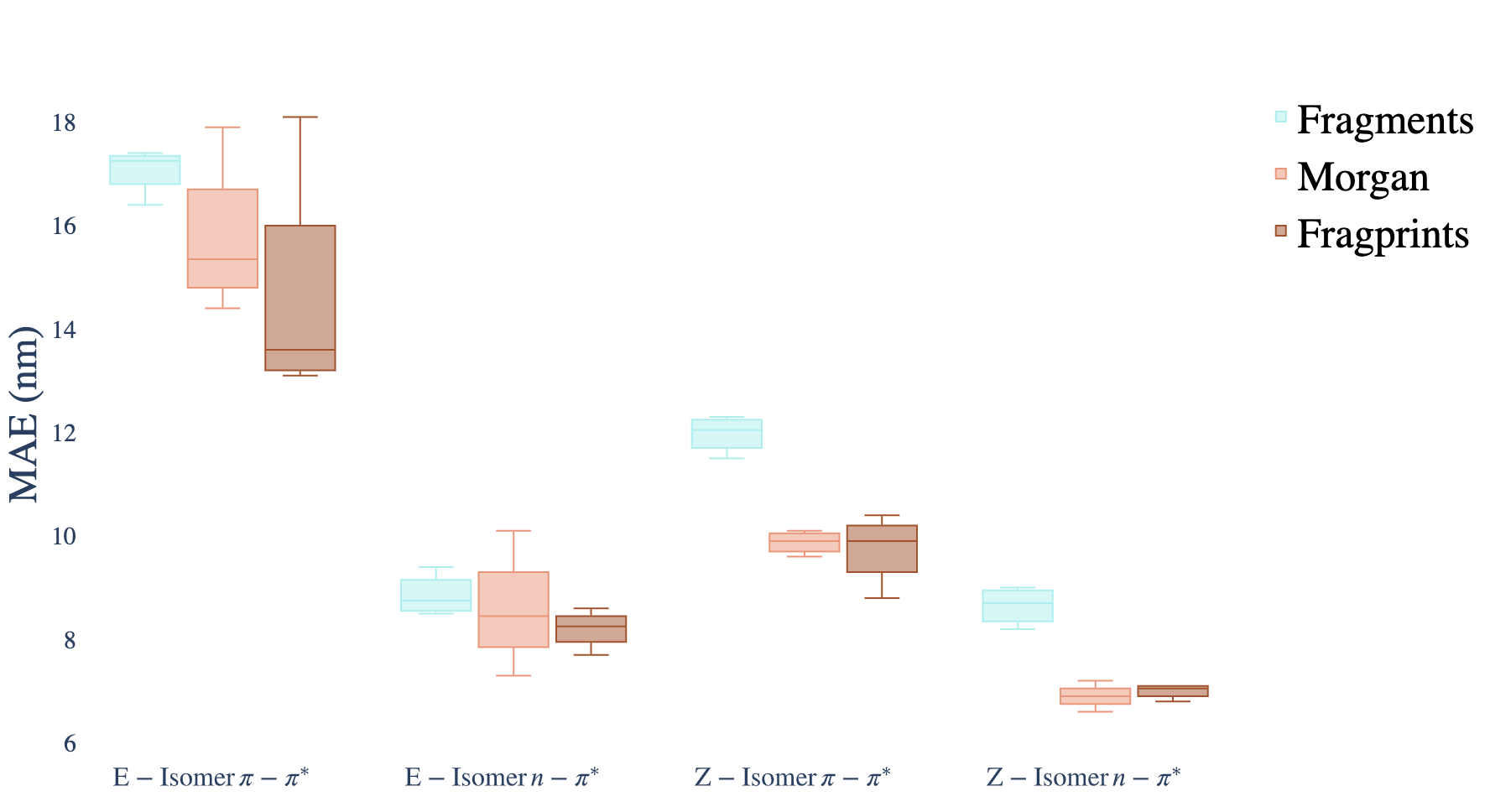
Prediction errors under different model/representation combinations may be analyzed in the context of the property being predicted.
The following boxplot shows shows the performance of each molecular representation aggregated across the Random Forest, Gaussian Process, Multioutput Gaussian Process and Attentive Neural Process models.
For the comparison with Time-Dependent Density Functional Theory (TD-DFT) run the DFT comparison script specifying the level of theory to compare against using the theory level flag (CAM-B3LYP or PBE0) e.g.
python dft_comparison_with_GPR.py -theory_level CAM-B3LYP | Metric | GPR-Tanimoto + Fragprints | CAM-B3LYP TD-DFT | CAM-B3LYP + Linear |
|---|---|---|---|
| MAE | 14.9 nm | 16.5 nm | 10.7 nm |
python dft_comparison_with_GPR.py -theory_level PBE0 | Metric | GPR-Tanimoto + Fragprints | PBE0 TD-DFT | PBE0 + Linear |
|---|---|---|---|
| MAE | 15.2 nm | 26.0 nm | 12.4 nm |
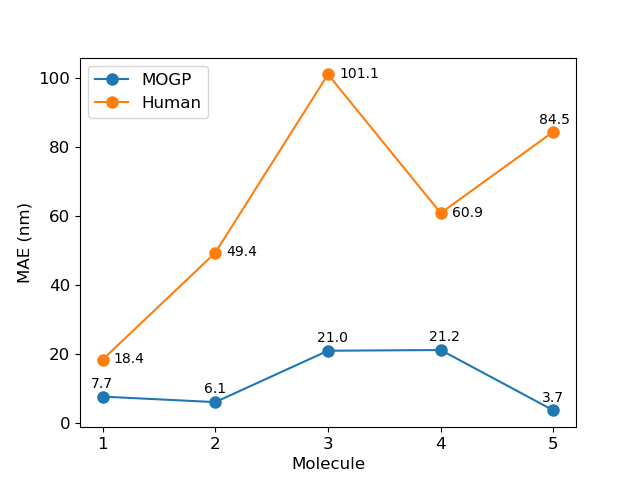
To reproduce the model prediction errors for the human performance comparison, run the following script:
python human_performance_comparison.py To reproduce the generalization error results, run the following scripts:
python generalization_error.py -augment_photo_dataset False | Metric | RF + Fragprints |
|---|---|
| RMSE | 85.2 nm |
| MAE | 72.5 nm |
| R2 | -0.66 |
python generalization_error.py -augment_photo_dataset True | Metric | RF + Fragprints |
|---|---|
| RMSE | 36.9 nm |
| MAE | 22.7 nm |
| R2 | 0.67 |
Run
python visualization.py to obtain an (unannotated) visualization of the Photoswitch Dataset
The dataset includes molecular properties for 405 photoswitch molecules. All molecular structures are denoted according to the simplified molecular input line entry system (SMILES). We collate the following properties for the molecules:
Rate of Thermal Isomerization (units = s^-1): This is a measure of the thermal stability of the least stable isomer (Z isomer for non-cyclic azophotoswitches and E isomer for cyclic azophotoswitches). Measurements are carried out in solution with the compounds dissolved in the stated solvents.
Photostationary State (units = % of stated isomer): Upon continuous irradiation of an azophotoswitch a steady state distribution of the E and Z isomers is achieved. Measurements are carried out in solution with the compounds dissolved in the ‘irradiation solvents’.
pi-pi-star/n-pi-star wavelength (units = nanometers): The wavelength at which the pi-pi*/n-pi* electronic transition has a maxima for the stated isomer. Measurements are carried out in solution with the compounds dissolved in the ‘irradiation solvents’.
DFT-computed pi-pi-star/n-pi-star wavelengths (units = nanometers): DFT-computed wavelengths at which the pi-pi*/n-pi* electronic transition has a maxima for the stated isomer.
Extinction coefficient: The molar extinction coefficient.
Wiberg Index: A measure of the bond order of the N=N bond in an azophotoswitch. Bond order is a measure of the ‘strength’ of said chemical bond. This value is computed theoretically.
Irradiation wavelength: The specific wavelength of light used to irradiate samples from E-Z or Z-E such that a photo stationary state is obtained. Measurements are carried out in solution with the compounds dissolved in the ‘irradiation solvents’.
Please see the examples folder for a tutorial on how to implement and use the GP-Tanimoto model in GPflow.
If you find the Photoswitch Dataset useful for your research, please consider citing the following article.
@article{2022Griffiths,
title={Data-driven discovery of molecular photoswitches with multioutput Gaussian processes},
author={Griffiths, Ryan-Rhys and Greenfield, Jake L and Thawani, Aditya R and Jamasb, Arian R and Moss, Henry B and Bourached, Anthony and Jones, Penelope and McCorkindale, William and Aldrick, Alexander A and Fuchter, Matthew J and others},
journal={Chemical Science},
volume={13},
number={45},
pages={13541--13551},
year={2022},
publisher={Royal Society of Chemistry}
}