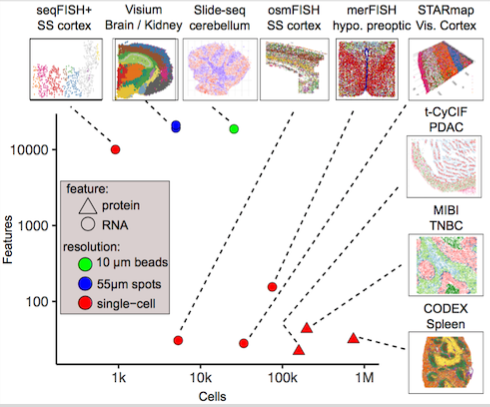
The Giotto package consists of two modules, Giotto Analyzer and Viewer (see www.spatialgiotto.com), which provide tools to process, analyze and visualize single-cell spatial expression data. The underlying framework is generalizable to virtually all currently available spatial datasets. We recently demonstrated the general applicability on 10 different datasets created by 9 different state-of-the-art spatial technologies, including in situ hybridization (seqFISH+, merFISH, osmFISH), sequencing (Slide-seq, Visium, STARmap) and imaging-based multiplexing/proteomics (CyCIF, MIBI, CODEX). These technologies differ in terms of resolution (single cell vs multiple cells), spatial dimension (2D vs 3D), molecular modality (protein vs RNA), and throughput (number of cells and genes). More information and documentation about the latest development version of Giotto Analyzer can be found at https://rubd.github.io/Giotto_site_dev/.