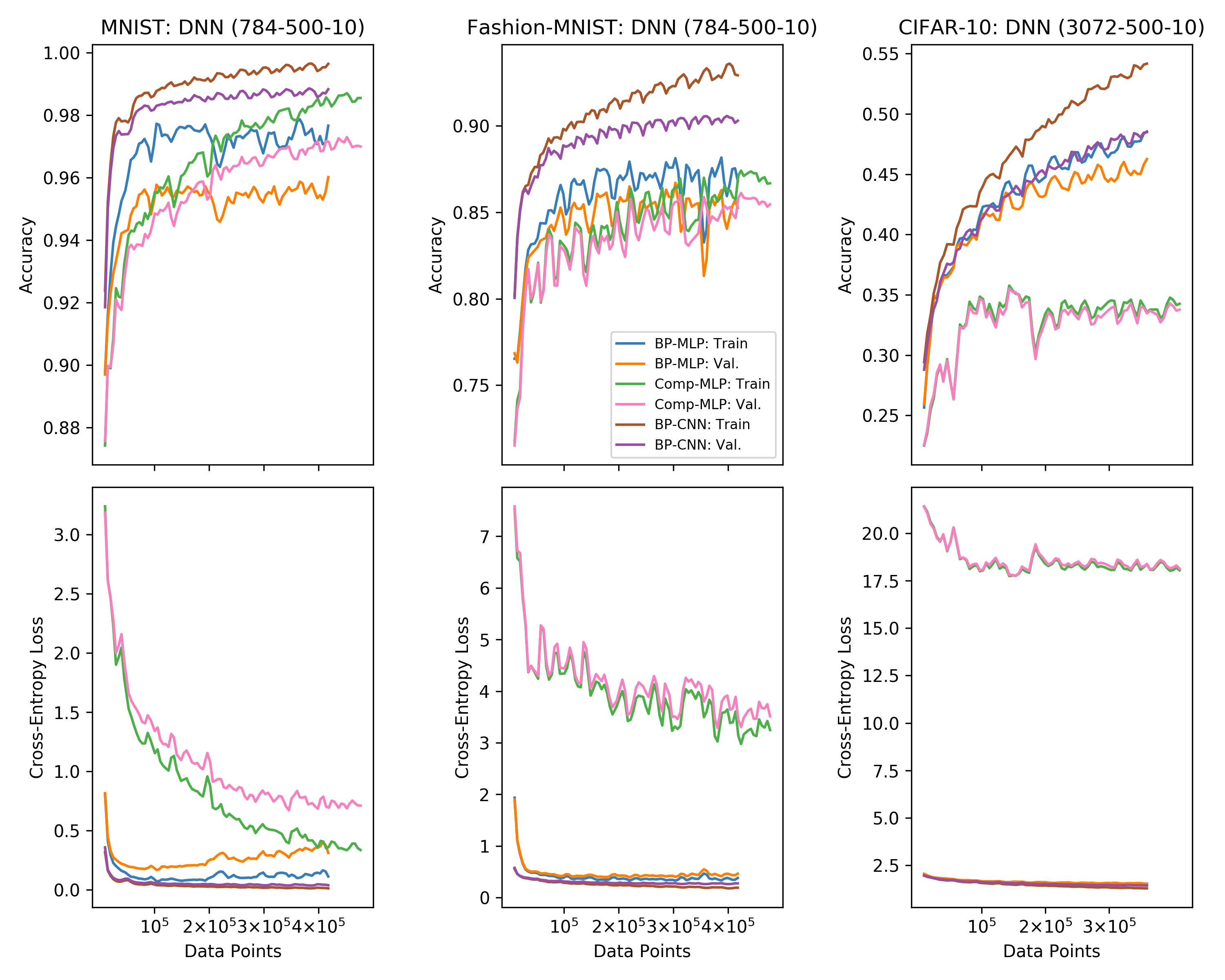
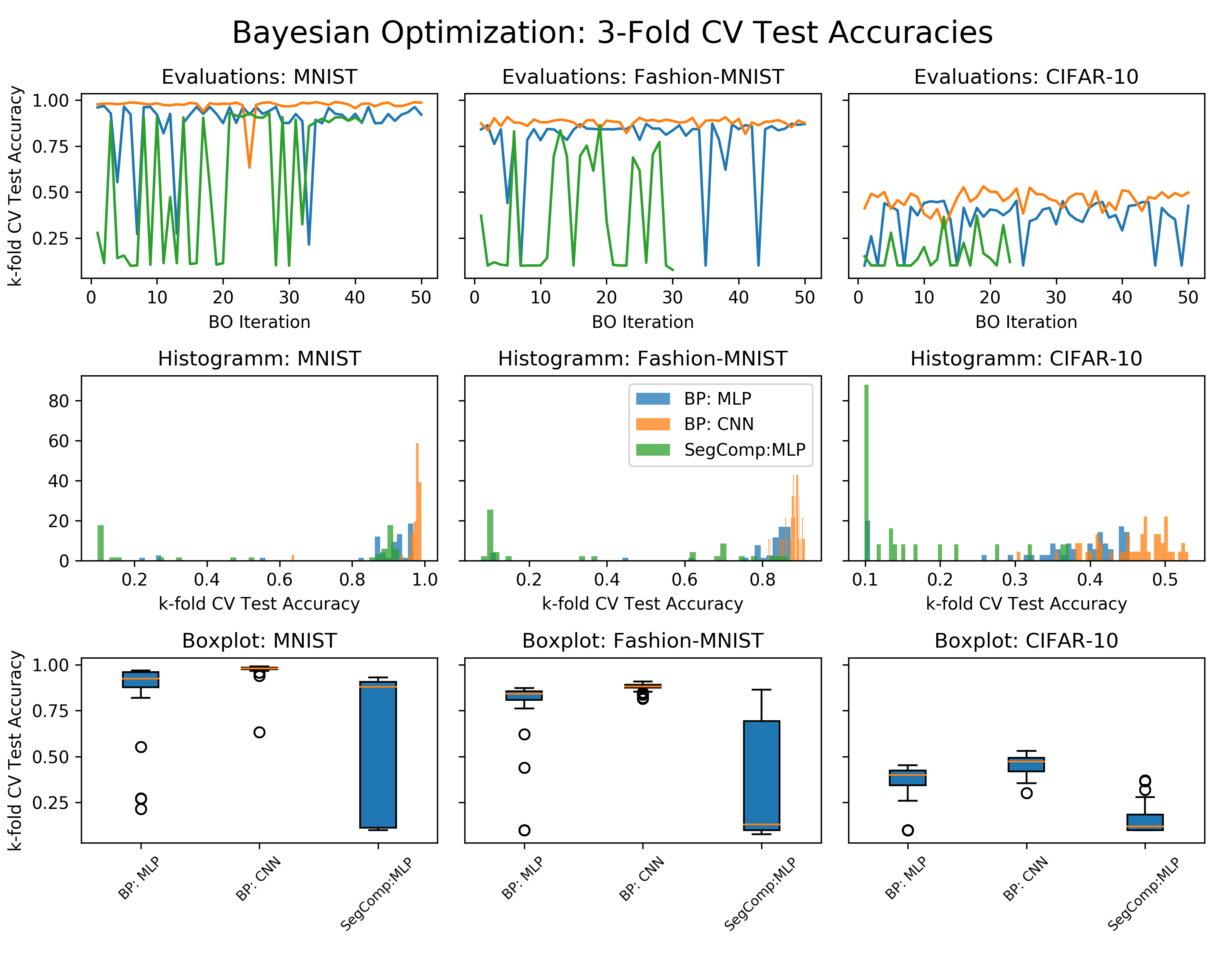
This project analyzes the performance and dynamics different learning rules in deep layered structures. More specifically, we explore alternatives to backpropagation (aka the chain rule). A few reasons for why this is interesting:
- Weight transport (access to all weights at every layer of the backward pass) renders backpropagation biologically implausible.
- Global signed error transmission as well as matrix transposition both are computationally expensive.
- The brain is more about long-range communication than computation.
Recent alternatives explore local learning rules and draw inspiration from the compartmental design of pyramidal neurons. Apical compartments integrate top-down information while basal dendritic compartments collect bottom-up information. This way one does not require separate pathways and instead exploits the electrical segregation observed in the physiology of pyramidal neurons in sensory cortices.
Here we reimplement Guerguiev, J., Lillicrap, T. P., & Richards, B. A. (2017). Towards deep learning with segregated dendrites. ELife, 6, e22901. and perform multiple robustness checks. Part of the code is adopted from their base implementation.
For more words please check out the following two documents:
Bio-Plausible-DeepLearning
├── workspace_dnn.ipynb: Trains individual backpropagation MLP/CNN models and runs Bayesian optimization pipeline for them.
├── workspace_comp_dnn.ipynb: Trains individual compartmental MLP models.
├── workspace_comp_visualize.ipynb: Produces the figures.
├── run_bo.py: Script for Bayesian Optimization.
├── figures: Folder containing saved figures.
├── logs: Folder containing training and optimization logs
├── models: Folder containing scripts defining DNN/CNN/CompDNN models
├── report: Folder containing writeup and presentation slides
├── utils: Helper functions (data preparation, logging, plotting, BO)
├── Readme.md: Documentation
├── requirements.txt: Dependencies
- Clone the repo.
git clone https://github.com/RobertTLange/Bio-Plausible-DeepLearning
cd Bio-Plausible-DeepLearning
- Create a virtual environment (optional but recommended).
virtualenv -p python BioDL
Activate the env (the following command works on Linux, other operating systems might differ):
source BioDL/bin/activate
- Install all dependencies:
pip install -r requirements.txt
- Run the main notebooks:
jupyter notebook workspace_dnn.ipynb
jupyter notebook workspace_guergiev.ipynb
- Run the bayesian optimization pipeline separately for compartmental DNN:
python run_bo.py -t comp_dnn -d mnist
python run_bo.py -t comp_dnn -d fashion
python run_bo.py -t comp_dnn -d cifar10
- Run the visualization notebook:
jupyter notebook workspace_visualize.ipynb
During the course of this project I trained many models. More specifically, I run 50 iterations of the BO pipeline for all three analyzed datasets as well as all three model types (Backprop MLP, Backprop CNN, Segregated Comp MLP). Running the Bayesian Optimization (BO) pipeline takes a while. Therefore, I run most of the analysis on a p2.xlarge AWS EC2 instance which utilizes a Tesla K80 GPU. In total training should take no more than 24 hours. Depending on the AMI that you use and whether or not you use a demand/spot instance, this should cost around 15$. I recommend the AWS Deep Learning Base AMI.
- Clone repo, Create/Activate the environment and install dependencies
git clone https://github.com/RobertTLange/Bio-Plausible-DeepLearning
cd Bio-Plausible-DeepLearning
conda create --name BioDL python=3.6 --no-default-packages
source activate BioDL
pip install -r requirements.txt --quiet
- Add ipykernel to listed env kernels, Launch notebook silent and open port (start a screen session in between!)
python -m ipykernel install --user --name BioDL --display-name "Python3 (BioDL)"
jupyter notebook --no-browser --port=8080
- In new terminal window on local machine rewire port and listen
ssh -i keyname.pem -N -f -L localhost:2411:localhost:8080 user@MACHINE_IP_ADDRESS
- In Browser open localhost port and start working on the notebook of choice. If required copy paste the token/set a password. Afterwards run notebook of choice
localhost:2411
- After computations are done either git add, comnmit, push to remote repo or copy files back.
scp -i keyname.pem -r user@MACHINE_IP_ADDRESS:Bio-Plausible-DeepLearning .
conda env remove -n BioDL
jupyter kernelspec uninstall BioDL