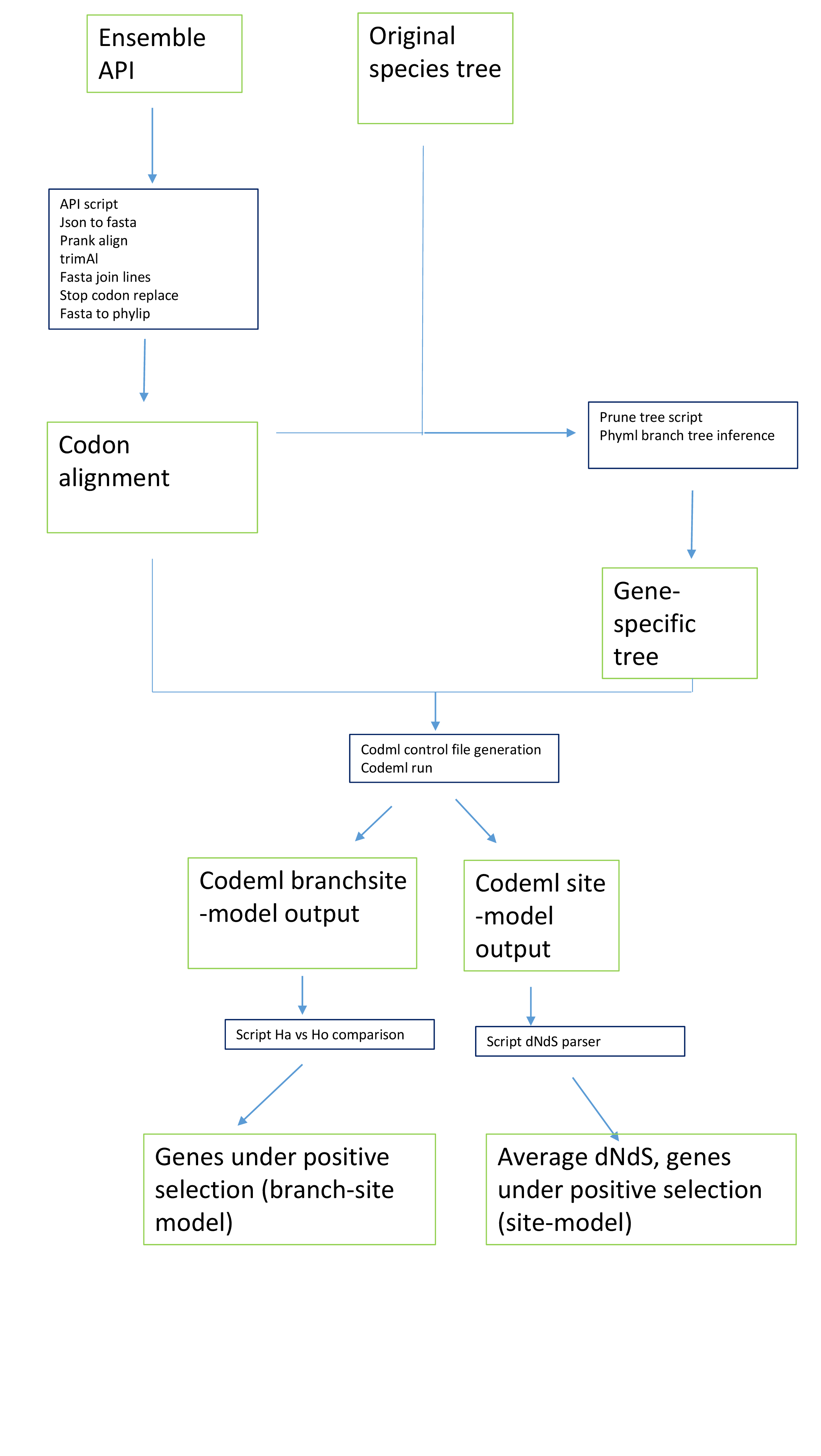
With this repository, we want to study the selective pressure of ~14000 human genes. Specifically, detect those genes under positive selection in humans compared to other primates, and estimate their dN/dS.
Contributors: Kernyu, Juan and Albert
Make the following pipeline
-
Install
jqfirst$bash src/ensembl_filter_json.sh CCR5 cdna | python src/json2fasta.py
$bash src/ensembl_filter_json.sh CCR5 protein | python src/json2fasta.py
- First Download the fasta files using Ensembl API
$bash src/ensembl_filter_json.sh <GENE_NAME> cdna | python src/json2fasta.py
- Do translation-aware alignment : Use
prankto do it
ls data/fasta/* | grep --file=20170921-list-genes-that-with-no-alignment.txt | parallel "/Users/akl2140/bin/prank/bin/prank -d={} -o=data/fasta-aligned/{/.}.aligned.fa -translate -F"
-
Remove gappy regions, using trimAl ("gappyout" settings)
-
Primate's tree prunning (scripts name?)
-
Estimate branch length for the prunned tree of each gene using PhyML. Fasta file has to be converted into phylip format first.
ls data/fasta/*fa | parallel --dry-run "Rscript src/fa2phyinter.R {/.} data/fasta data/phylip && phyml -i data/phylip/{/.}.phy -d nt -b 0 -m GTR -c 4 -a 1 -u data/pruned_tree/{/.}.tree -o lr" | bash
- Run codeml (branch-site or site model). Input files: i) sequene alignment (phylip format), ii) gene specific tree, iii) contro (ctl) file for codeml. We have uploaded bash scripts to generate the ctl files for either codeml model (assets folder).
# Rscript src/codeml-process-pvalue.R <NULL MODEL MLC FILE> <ALTERNATIVE MODEL MLC FILE>
Rscript src/codeml-process-pvalue.R data/paml_results/A1BG_H0.mlc data/paml_results/A1BG_HA.mlc
Rscript src/codeml-process-table-site-class.R <MLC FILE> # will produce csv output