Documentation for running ld-score regression between GWAS summary statistics from a reference phenotype and set of phenotypes of interest.
- Unix-based operating system (Linux or OSx. Sorry Windows folks.)
- Java v1.8 and higher (download here)
- Docker
- AWS CLI
-
Install Cromwell if you haven't already
-
Configure the AWS CLI for use with CODE AWS group.
- Configure with the secret key associated with the CODE AWS account
-
Clone local copy of ld-regression-pipeline to download this repository to your local machine
git clone https://github.com/RTIInternational/ld-regression-pipeline.git
Voila! You're ready to get started running the pipeline.
More detailed running instructions for WDL/Cromwell workflows are found in the tutorial for the metaxcan pipeline here
LD-Hub and the associated LDSC software package provides a suite of tools designed to perform LD-score regression between GWAS outputs of two phenotypes.
This document describes an automated WDL workflow designed to perform LD regression of a reference phenotype against one or more phenotypes using the following high level steps:
- Transform GWAS summary statistics into summary statistics format
- Reformat summary statistics using
munge_sumstats.py - Perform LD-regression between the reference and all traits of interest using
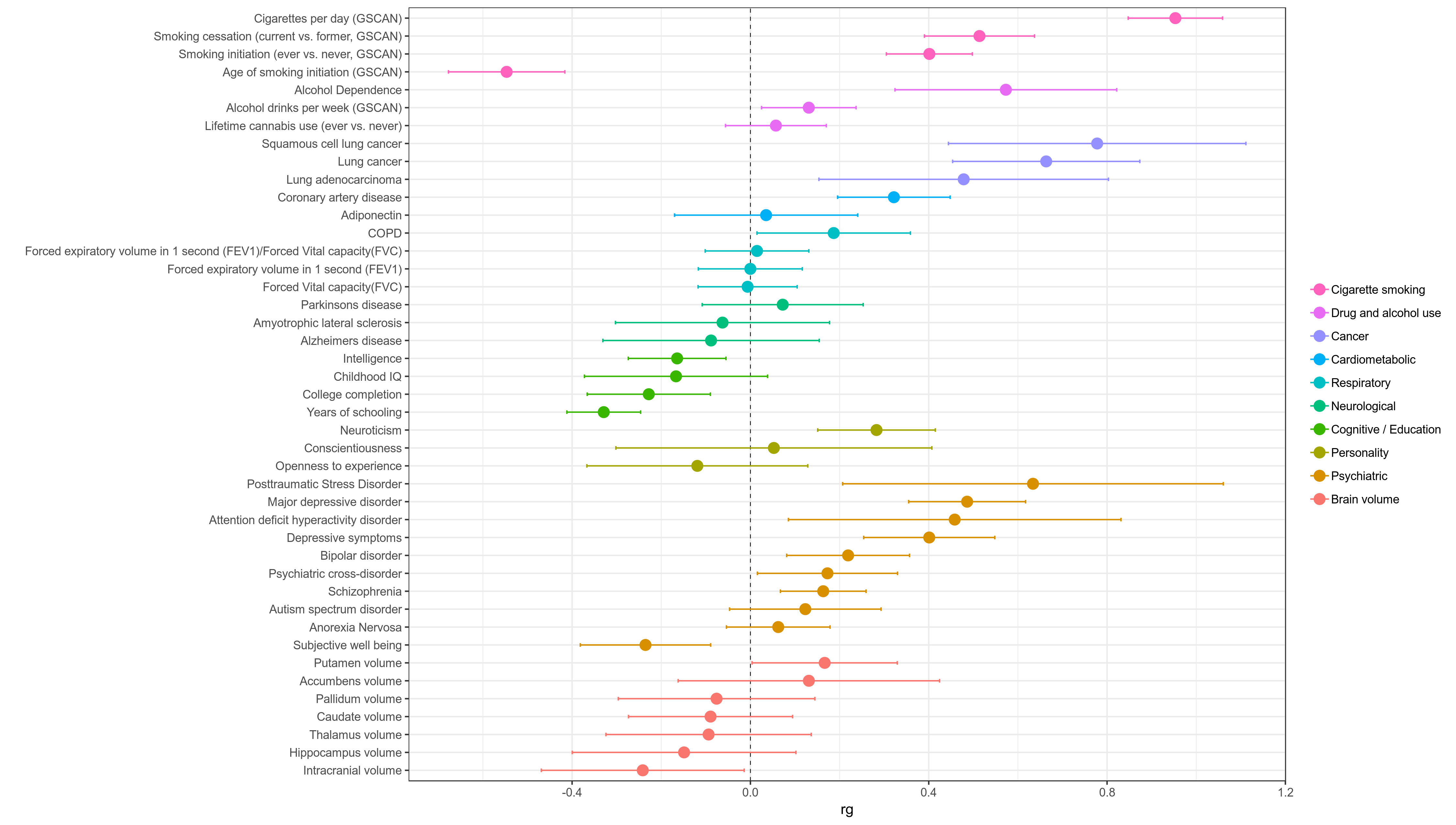
ldsc.py - Combine results and plot in a single graph (below)
The following are required for the main phenotype and each phenotype you want to compare using ld-regression:
- Phenotype name
- Plot label for how you want the phenotype to be labeled on the output graph
- path to a GWAS summary statistics output file
- 1-based column index for each of the following in the GWAS input file:
- id col
- chr col
- pos col
- effect allele col
- ref allele col
- pvalue col
- Effect column (e.g. beta, z-score, odds ratio)
- Either the number of samples in the cohort or the sample size column index in the GWAS input
- Effect column type in the GWAS file (BETA, Z, OR, LOG_ODD)
- Plot group for grouping phenotypes into larger trait groups on final output plot
- Transform GWAS in summary statistics
- Split by chr if necessary
- Put columns in standardized order and give standard colnames (e.g. SNP, A1, A2, BETA, etc.)
- Convert to 1000g ids
- Replace 1000g ids with only rsIDs
- Split by chr if necessary
- Reformat summary statistics using
munge_sumstats.py- For each chr:
- Call munge_sumstats.py
- Remove empty variants for each chromosome
- Re-combine into single munged sumstats file
- For each chr:
- Perform LD-regression between the reference and all traits of interest using
ldsc.py- For each phenotype you want to regress against reference phenotype
- untar ld reference panel files
- Call
LDSC.pywith--rgoption to get logfile with regression statistics
- For each phenotype you want to regress against reference phenotype
- Combine results and plot in a single graph
- Parse statistics from log files generated by LDSC
- Combine into single table and add plot information (e.g. plot_label, plot_group)
- Make plot of rg values returned for each phenotype with 95% CI error bars
This repo provides a custom docker tool designed to help generate the json-input file needed to run the pipeline. To generate your analysis-ready input you need to:
- Fill out your workflow template file
- Create a pheno info excel spreadsheet containing info about phenotypes you want to compare
- Use generate_ld_regression_input_json.py tool to create your final input file
Start with the json_input/full_ld_regression_wf_template.json and fill in the following values unique to your analysis:
-
Give your analysis a name
"full_ld_regression_wf.analysis_name": "ftnd_test" -
Set the name of your ref trait.
"full_ld_regression_wf.ref_trait_name" : "ftnd", -
Specify paths to GWAS summary files (22 in total; 1 per chr) for your main reference trait you wish to compare to other phenotypes
"full_ld_regression_wf.ref_sumstats_files": [ "s3://rti-nd/META/1df/boneyard/20181108/results/ea/20181108_ftnd_meta_analysis_wave3.eur.chr1.exclude_singletons.1df.gz", "s3://rti-nd/META/1df/boneyard/20181108/results/ea/20181108_ftnd_meta_analysis_wave3.eur.chr2.exclude_singletons.1df.gz", ... "s3://rti-nd/META/1df/boneyard/20181108/results/ea/20181108_ftnd_meta_analysis_wave3.eur.chr22.exclude_singletons.1df.gz", ] -
Inspect your GWAS summary files to find the correct indices for each required column index (1-based so the first column has index 1)
"full_ld_regression_wf.ref_id_col": 1, "full_ld_regression_wf.ref_chr_col": 2, "full_ld_regression_wf.ref_pos_col": 3, "full_ld_regression_wf.ref_effect_allele_col": 4, "full_ld_regression_wf.ref_ref_allele_col": 5, "full_ld_regression_wf.ref_beta_col": 6, "full_ld_regression_wf.ref_pvalue_col": 8, -
If your GWAS summary stats file has a sample size column, specify it's index
"full_ld_regression_wf.ref_num_samples_col": 11 -
Otherwise, provide the number of samples in the cohort (will be the same for all SNPs)
"full_ld_regression_wf.ref_num_samples" : 45233 -
Specify the effect type and the zero value as you would pass it to
munge_sumstats.py"full_ld_regression_wf.ref_signed_sumstats" : "BETA,0" -
Provide paths to the reference ld-reference panel bz2 tarball passed to
LDSC.py"full_ld_regression_wf.ref_ld_chr_tarfile": "s3://clustername--files/eur_w_ld_chr.tar.bz2", -
Provide path to a merge_allele snplist needed for
munge_sumstats.py"full_ld_regression_wf.merge_allele_snplist": "s3://clustername--files/w_hm3.snplist",
Example pheno input file here
The pheno info excel spreadsheet contains information for each phenotype to compare per row.
The format requires the following column names (explanation in parentheses):
- trait (Trait Name)
- plot_label (Label to be used on plots)
- sumstats_path (Path to GWAS summary stats file)
- category (Group label for plotting)
- sample_size (GWAS sample size)
- effect_type (Effect type; accepts Beta, Z, OR, log_odd)
- w_ld_chr (Path to LD reference tarball to be used for
LDSC.py) - id_col (1-based column index of snp id in GWAS sumstats file)
- chr_col (chr column index)
- pos_col (position index)
- effect_allele_col (effect allele index)
- ref_allele_col (ref allele index)
- effect_col (effect score index)
- pvalue_col (pvalue index)
- sample_size_col (sample size column index if present)
Note: All phenotypes must have either sample_size or sample_size_col but do not need to have both.
Create a folder that contains the input template and your phenotype file
mkdir /path/to/workflow_inputs
cp json_input/full_ld_regression_wf_template.json workflow_inputs
cp phenotype_file.xlsx workflow inputs
Run inside docker image to produce final input file (Note: output gets printed to stdout)
docker run -v "/path/to/workflow_inputs:/data/" rticode/generate_ld_regression_input_json \
--json-input /data/full_ld_regression_wf_template.json \
--pheno-file /data/phenotype_file.xlsx > /data/final_wf_inputs.json
Now the analysis-ready input file is ready to run and will be located at /path/to/workflow_inputs/final_wf_inputs.json
First, make a zip file of the git repo you cloned so Cromwell can handle the local WDL imports:
# Change to directory immediately above metaxcan-pipeline repo
cd /path/to/ld-regression-pipeline
cd ..
# Make zipped copy of repo somewhere
zip --exclude=*var/* --exclude=*.git/* -r /path/to/workflow_inputs/ld-regression-pipeline.zip ld-regression-pipeline
Now run either via command line:
java -Dconfig.file=/path/to/aws.conf -jar cromwell-36.jar run \
workflow/plot_ld_regression_wf.wdl \
-i /path/to/workflow_inputs/final_wf_inputs.json \
-p /path/to/workflow_inputs/ld-regression-pipeline.zip
Or via an API call to a Cromwell server on AWS:
curl -X POST "http://localhost:8000/api/workflows/v1" -H "accept: application/json" \
-F "workflowSource=@/full/path/to/workflow/full_ld_regression_wf.wdl" \
-F "workflowInputs=@"/path/to/workflow_inputs/final_wf_inputs.json \
-F "workflowDependencies"=@/path/to/workflow_inputs/ld-regression-pipeline.zip
There are a number of workflows if you're interested in running only part of the pipeline manually:
- ldsc_preprocessing_wf.wdl: run the pipeline but stop after
munge_sumstats.py - munge_phenotype_sumstats_wf.wdl: run a single GWAS sumstats file through the
munge_sumstats.pypipeline - munge_sumstats_wf.wdl: run a single GWAS sumstats file that has already been split by chr through the
munge_sumstats.pypipeline - plot_ld_regression_wf.wdl: Merge and plot results from one or more ld-regression analyses
- single_ld_regression_wf.wdl: Run LD-regression only of the ref trait against a single trait
For any questions, comments, concerns, or bugs, send me an email or slack and I'll be happy to help.