If you use this work in published research, please cite:
G Yu*. Using ggtree to Visualize Data on Tree-Like Structure. Current Protocols in Bioinformatics, 2020, 69(1):e96, https://doi.org/10.1002/cpbi.96.
This repo contains source code and data to reproduce figures of the above paper.
library(treeio)
library(ggtree)
treetext = "(((ADH2:0.1[&&NHX:S=human], ADH1:0.11[&&NHX:S=human]):
0.05 [&&NHX:S=primates:D=Y:B=100],ADHY:
0.1[&&NHX:S=nematode],ADHX:0.12 [&&NHX:S=insect]):
0.1[&&NHX:S=metazoa:D=N],(ADH4:0.09[&&NHX:S=yeast],
ADH3:0.13[&&NHX:S=yeast], ADH2:0.12[&&NHX:S=yeast],
ADH1:0.11[&&NHX:S=yeast]):0.1[&&NHX:S=Fungi])[&&NHX:D=N];"
tree <- read.nhx(textConnection(treetext))
p <- ggtree(tree)
p2 <- p + geom_tiplab() +
geom_label(aes(x=branch, label=S), fill='lightgreen') +
geom_label(aes(label=D), fill='steelblue') +
geom_text(aes(label=B), hjust=-.5) +
xlim(NA, 0.28)
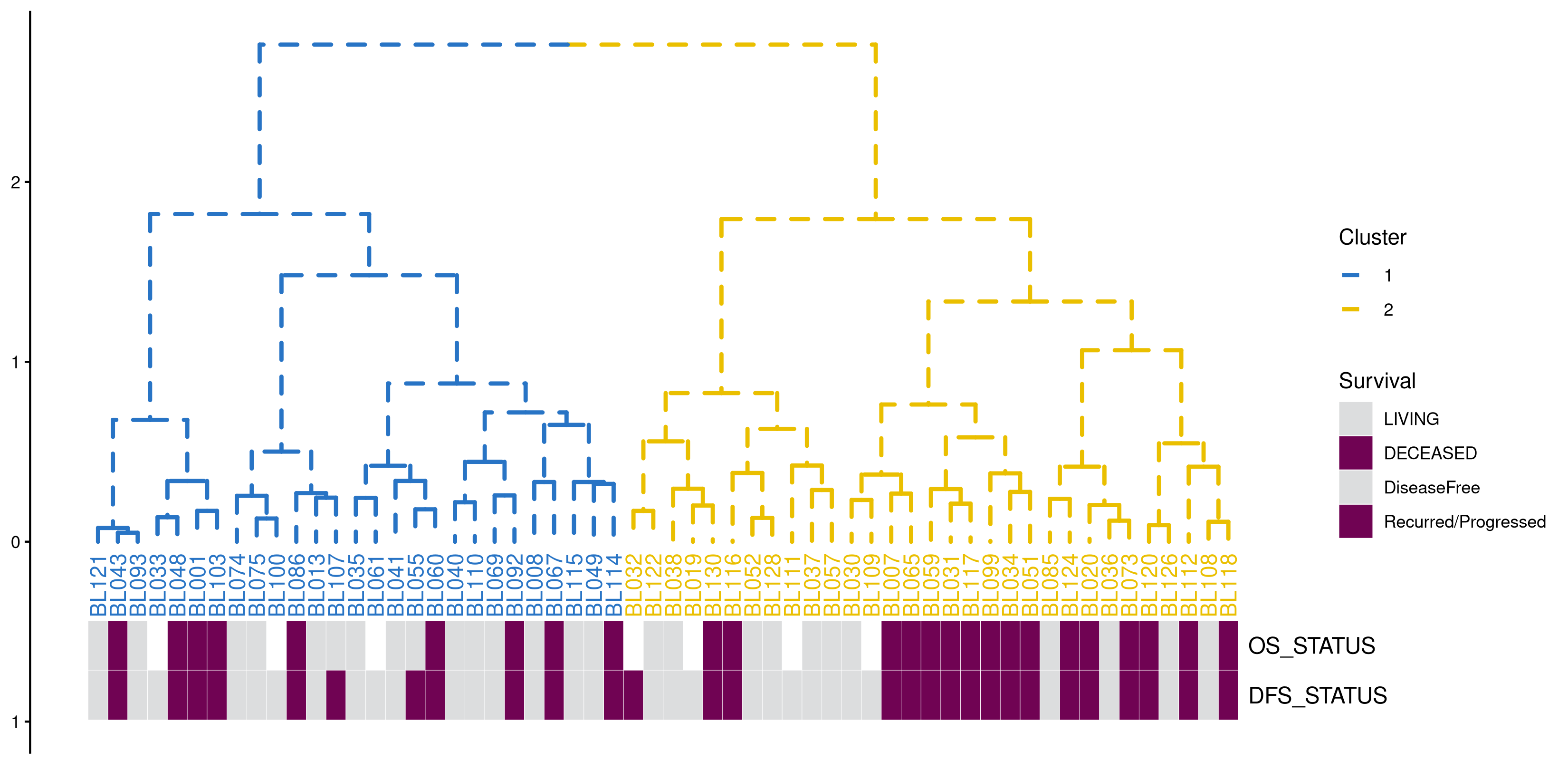
expr <- read.table("mskcc.txt")
d <- as.dist(1 - cor(expr, method="pearson"))
hc <- hclust(d, "ward.D")
p <- ggtree(hc, linetype='dashed',size=1) + layout_dendrogram() +
theme_dendrogram() +
geom_tiplab(angle=90, hjust=1, offset=-.05, show.legend=FALSE)
clus <- cutree(hc,k=2)
g <- split(names(clus), clus)
p <- groupOTU(p, g, group_name='Cluster') + aes(color=Cluster) +
scale_color_manual(breaks=c(1,2), values=c("#2874C5","#EABF00"))
surv <- read.table("mskcc_surv.txt")
surv.df <- data.frame(label = names(clus),
OS=surv$OS_STATUS,
PFS=surv$DFS_STATUS)
g2 <- p %<+% surv.df +
geom_tippoint(shape=21, aes(fill=OS), color='black', size=6) +
scale_fill_manual(values=c("LIVING"="#dcddde","DECEASED"="#700353"),
na.value="white") +
ggnewscale::new_scale_fill() +
geom_tippoint(shape=21, aes(fill=PFS), color='black', size=3) +
scale_fill_manual(values=c("DiseaseFree"="#dcddde",
"Recurred/Progressed"="#700353")) +
theme(plot.margin=margin(6,6,30,6), legend.position=c(.5, .6))
g3 <- gheatmap(p, surv[, c("OS_STATUS", "DFS_STATUS")], width=.2,
colnames_position="top", offset=.3, hjust=0) +
scale_fill_manual(breaks=c("LIVING", "DECEASED",
"DiseaseFree", "Recurred/Progressed"),
values=c('#700353', '#dcddde', '#dcddde', '#700353'),
na.value="white", name = "Survival")
## Scale for 'fill' is already present. Adding another scale for 'fill', which
## will replace the existing scale.
library(ape)
library(ggplot2)
library(tidytree)
library(treeio)
library(ggtree)
data(woodmouse)
d <- dist.dna(woodmouse)
tr <- nj(d)
bp <- boot.phylo(tr, woodmouse, function(x) nj(dist.dna(x)))
##
Running bootstraps: 100 / 100
## Calculating bootstrap values... done.
bp2 <- tibble(node=1:Nnode(tr) + Ntip(tr), bootstrap = bp)
tree <- full_join(tr, bp2, by="node")
g4 = ggtree(tree, size=1.5) +
geom_tiplab(size=4.5, fontface="bold") + xlim(0, 0.02) +
geom_nodepoint(aes(fill=cut(bootstrap, c(0, 70, 90, 100))),
shape=21, size=4) +
theme_tree(legend.position=c(0.8, 0.2)) +
scale_fill_manual(values=c("white", "grey", "black"), guide='legend',
name='Bootstrap Percentage(BP)',
breaks=c('(90,100]', '(70,90]', '(0,70]'),
labels=expression(BP>=90,70 <= BP * " < 90", BP < 70))
library(ggimage)
library(ggtree)
tree <- read.tree("tree_boots.nwk")
info <- read.csv("taxa_info.csv")
mass <- info$mass_in_kg
names(mass) <- info$Newick_label
fit <- phytools::fastAnc(tree,mass,vars=TRUE,CI=TRUE)
td <- data.frame(node = nodeid(tree, names(mass)),
trait = mass)
nd <- data.frame(node = as.numeric(names(fit$ace)),
trait = fit$ace)
d <- rbind(td, nd)
library(treeio)
tree2 <- full_join(tree, d, by = 'node')
pg <- ggtree(tree2, aes(color=trait), continuous = TRUE, size=3) +
scale_color_gradientn(colours=c("red", 'orange', 'green', 'cyan', 'blue'),
name="mass (kg)") +
geom_tiplab(hjust = -.2) + xlim(0, 4)
trophic_habit <- setNames(info$trophic_habit, info$Newick_label)
cols <- RColorBrewer::brewer.pal(length(unique(trophic_habit)), "Set1")
names(cols) <- sort(unique(trophic_habit))
fitER <- ape::ace(trophic_habit,tree,model="ER",type="discrete")
ancstats <- as.data.frame(fitER$lik.anc)
ancstats$node <- 1:Nnode(tree)+Ntip(tree)
pies <- nodepie(ancstats, cols=1:3)
pies <- lapply(pies, function(g) g + scale_fill_manual(values = cols))
pg <- pg %<+% info +
geom_tippoint(aes(fill = trophic_habit), shape=21, size=10, color='white') +
scale_fill_manual(values = cols) +
geom_inset(pies, width = .2, height=.2)
id <- phylopic_uid(info[,1])
gg <- pg %<+% id +
geom_tiplab(aes(image=uid), geom="phylopic", offset=.75, size=.1)
library(ape)
library(Biostrings)
fasfile <- system.file("extdata/sample.fasta", package = "ggmsa")
x <- readAAStringSet(fasfile)
d <- as.dist(stringDist(x, method = "hamming")/width(x)[1])
tree <- bionj(d)
library(ggtree)
library(ggmsa)
p <- ggtree(tree, size=1) + geom_tiplab()
data <- tidy_msa(fasfile, start = 160, end = 220)
p2 <- p +
geom_facet(geom = geom_msa, data = data,
panel = 'Multiple Sequence Alignment',
color = "Chemistry_AA") +
xlim_tree(3)
pp <- facet_widths(p2, widths=c(.2, 1))