Hierarchical SEed Aligner
HISEA is an efficient all-vs-all long read aligner for SMRT sequencing data. Its algorithm is designed to produce highest alignment sensitivity among others.
Further, HISEA is integrated in Canu assembly pipeline which primarily uses MHAP. The HISEA aligner produces better assembly than MHAP when used in Canu. The Canu+HISEA pipeline can be downloaded from here.
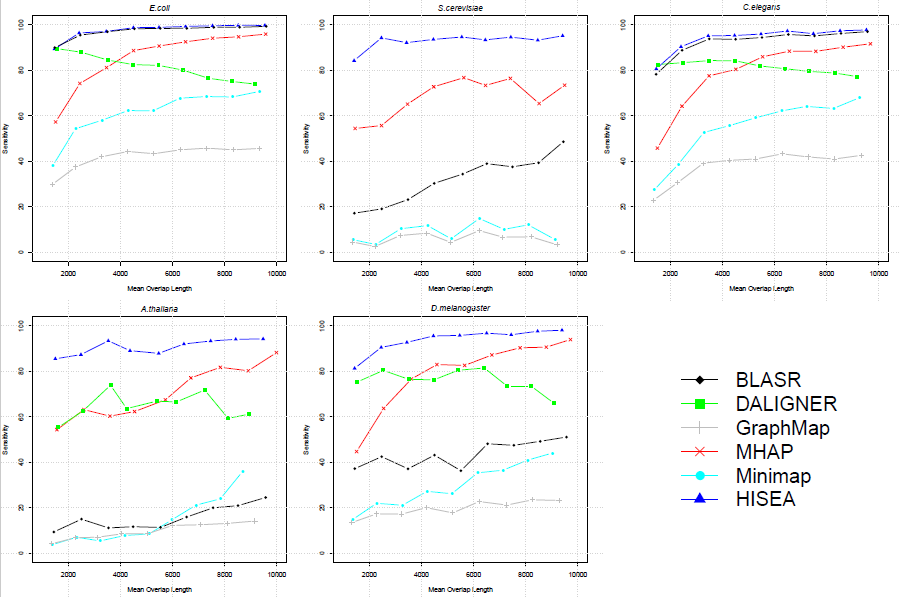
A detailed comparison of HISEA with other leading programs can be found in HISEA paper (Nilesh Khiste and Lucian Ilie). Below are some plots showing different comparisons.
We have used a modified version of EstimateROC utility from MHAP, for evaluating Sensitivity, Specificity and Precision. The modified EstimateROC Java file can be downloaded from here. To use modified EstimateROC utility, please replace your exiting EstimateROC.java file in MHAP with this copy and recompile MHAP code.
 |
 |
|---|---|
 |
 |
 |
It is expected that larger overlaps are easier to find compared to short ones. This comparison shows how different program behave with increasing overlap length. Clearly HISEA performs best irrespective of the overlap size.
Once the zip file has been downloaded, please use following commands to unzip and compile the HISEA program:
1. unzip HISEA.zip -- This will create a directory HISEA and all required files in this folder
2. cd HISEA -- Change directory to HISEA
3. make -- This will compile the HISEA code and create hisea file for useThis command will build the hisea binary. If you see any error messages, please contact authors.
The HISEA program can be used with fastA/fastQ files with one or more sequences. The program can be run in both serial and parallel mode. The parallel mode has an advantage in terms of time with respect to serial mode. HISEA can be run with minimal set of mandatory parameters, --ref and --query. For self alignment, HISEA only needs two options --self and --ref. All other parameter will assume their default values.
USAGE:
hisea [--self] [--kmerLen <int>] [--minStoreLen <int>] [--minOlapLen <int>] --ref <file/directory> --query <file/directory>
For full list of options type "hisea --help"
OUTPUT:
stdout List of alignmentsThe complete set of options:
Options:
--help no_arg Print this help message
--self no_arg Align set of reads with itself. Use only --ref option
--ref <file/dir> The name of the reference fasta/fastq file or directory
containing these files
--query <file/dir> The name of the query fasta/fastq file or directory
containing these files
--ignore <file> The file containing kmers to be ignored
--index_write <dir> The directory where index is stored
--index_read <dir> The directory from whcih index is read
--kmerLen <int> This is the kmer length used for initial hashing.
The possible values are 10-20, both inclusive
default=16
--smallkmerLen <int> This is the kmer length used during alignment extension.
The possible values are 10-20, both inclusive
default=12
--filterCount <int> This is used for initial filtering. This must be set to 1,
if index is created with split set of reads.
default=2
--threads <int> Number of threads for parallel run
default=1
--minOlapLen <int> Minimum Overlap length
default=100
--minStoreLen <int> Minimum read length. Overlap of two smaller reads is ignored
default=100
--minMatches <int> Minimum number of matches to be considered for alignment
default=3
--maxKmerHits <int> The maximum number of repeat kmers
default=10000
--errorRate <float> Error rate. Valid values are 0-1 percent
default=0.15
--maxShift <float> The value of shift to accomodate indels. Valid values
are 0-1 percent
default=0.20If you find HISEA program useful, please cite the HISEA paper:
N. Khiste, L. Ilie HISEA: HIerarchical SEed Aligner for PacBio data BMC Bioinformatics, 2017