longmethyl
Learning Nextflow - A demo nextflow pipeline of methylation detection using long reads
Contents
Installation
-
(1) Install conda from Conda if neeeded.
-
(2) Install nextflow.
# create a new environment and install nextflow in it
conda create -n nextflow -c conda-forge -c bioconda nextflow
# or install nextflow in an existing environment
conda install -c conda-forge -c bioconda nextflow- (3) Download longmethyl from github.
git clone https://github.com/PengNi/longmethyl.git-
(4) Install Docker or Singularity if needed.
-
(5) [optional] Install graphviz.
conda install -c conda-forge graphvizDemo data
Check longmethyl/demo for demo data:
- fast5_chr20.tar.gz: 60 HG002 fast5s which align to human genome chr20:10000000-10100000.
- chr20_demo.fa: reference sequence of human chr20:10000000-10100000.
- hg002_bsseq_chr20_demo.bed: HG002 BS-seq results of region chr20:10000000-10100000.
If you are using Conda to run longmethyl, check also google drive to get deepsignal CpG model-model.CpG.R9.4_1D.human_hx1.bn17.sn360.v0.1.7+.tar.gz.
Usage
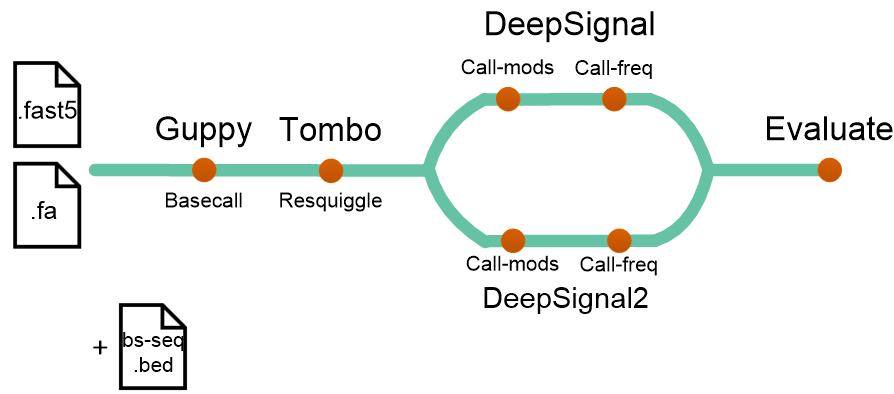
The longmethyl pipeline is for methylation calling from nanopore reads as following:
Option 1. Run with singularity (recommended)
If it is the first time you run with singularity (e.g. using -profile singularity), the following cmd will cache the dafault singularity image (--singularity_name) to the --singularity_cache directory (default: local_singularity_cache) first. There will be a .img file in the --singularity_cache directory.
# activate nextflow environment
conda activate nextflow
# run longmethyl, this cmd will cache a singularity image before processing the data
nextflow run ~/tools/longmethyl -profile singularity \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gz
# or, run longmethyl using GPU, set CUDA_VISIBLE_DEVICES
CUDA_VISIBLE_DEVICES=0 nextflow run ~/tools/longmethyl -profile singularity \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gzThe downloaded .img file can be used then, without being downloaded again:
# this time nextflow will not download the singularity image again, it has already
# been in the --singularity_cache directory.
nextflow run ~/tools/longmethyl -profile singularity \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gz
# or
nextflow run ~/tools/longmethyl -profile singularity \
--singularity_cache local_singularity_cache \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gz
# or
nextflow run ~/tools/longmethyl -profile singularity \
--singularity_name local_singularity_cache/nipengcsu-longmethyl-0.3.img \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gzThe singularity image can be also pulled before running the cmd. The pulled .sif file is only needed to be downloaded once.
# pull singularity image (once for all). There will be a .sif file.
singularity pull docker://nipengcsu/longmethyl:0.3
# run longmethyl
nextflow run ~/tools/longmethyl -profile singularity \
--singularity_name longmethyl_0.3.sif \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gzOption 2. Run with docker
- (1) Pull docker image (once for all).
It is better to pull docker image before running pipeline the first time, cause this may be time-consuming and there may be network issues. However, this step is not necessary, the image will be pulled automatically when running the pipeline the first time.
docker pull nipengcsu/longmethyl:0.3- (2) Run longmethyl using
-profile docker.
# activate nextflow environment
conda activate nextflow
# run longmethyl using cpu
nextflow run ~/tools/longmethyl -profile docker \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gzCurrently longmethyl CANNOT run with docker on a GPU machine.
# TODO: run longmethyl using GPU, set CUDA_VISIBLE_DEVICES and --gpu
CUDA_VISIBLE_DEVICES=0 nextflow run ~/tools/longmethyl -profile docker --gpu true \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gzRelated issues:
- For
No swap limit support
# for Ubuntu
# (1) sudo, Edit the /etc/default/grub file. Add or edit the GRUB_CMDLINE_LINUX line
# to add the following two key-value pairs
GRUB_CMDLINE_LINUX="cgroup_enable=memory swapaccount=1"
# (2) Update GRUB
sudo update-grub
# (3) Restart the machine
sudo rebootRef: https://unix.stackexchange.com/questions/342735/docker-warning-no-swap-limit-support
- For
docker: Error response from daemon: could not select device driver "" with capabilities: [[gpu]].
# for Ubuntu
distribution=$(. /etc/os-release;echo $ID$VERSION_ID) \
&& curl -s -L https://nvidia.github.io/nvidia-docker/gpgkey | sudo apt-key add - \
&& curl -s -L https://nvidia.github.io/nvidia-docker/$distribution/nvidia-docker.list | sudo tee /etc/apt/sources.list.d/nvidia-docker.list
sudo apt-get update
sudo apt-get install -y nvidia-docker2
sudo systemctl restart dockerRef: https://docs.nvidia.com/datacenter/cloud-native/container-toolkit/install-guide.html
- For
Failed to initialize NVML: Driver/library version mismatch
Option 3. Run with conda
- (1) Install the conda environment named longmethyl (once for all).
# in a gpu machine, make sure there is already cuda10.0 and cuda driver in the machine
conda env create -f longmethyl/environment.yml
# or, in a cpu-only machine
conda env create -f longmethyl/environment-cpu.yml-
(2) Install Guppy, since Guppy is not open-sourced, from ONT community (once for all).
-
(3) Download the pre-trained model of deepsignal for calling mods [check deepsignal CpG model-model.CpG.R9.4_1D.human_hx1.bn17.sn360.v0.1.7+.tar.gz in google drive].
-
(4) Run longmethyl using
-profile condaand the longmethyl environment.
# activate nextflow environment
conda activate nextflow
# run longmethyl
nextflow run ~/tools/longmethyl -profile conda \
--conda_name /home/nipeng/tools/miniconda3/envs/longmethyl \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gz \
--deepsignalDir model.CpG.R9.4_1D.human_hx1.bn17.sn360.v0.1.7+.tar.gz
# or, run longmethyl using GPU, set CUDA_VISIBLE_DEVICES
CUDA_VISIBLE_DEVICES=0 nextflow run ~/tools/longmethyl -profile conda \
--conda_name /home/nipeng/tools/miniconda3/envs/longmethyl \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gz \
--deepsignalDir model.CpG.R9.4_1D.human_hx1.bn17.sn360.v0.1.7+.tar.gzExtra 1. Run longmethyl and the benchmark process
If you want benchmark the ONT 5mCpG calling pipeline with something like BS-seq, set --eval_methcall as true and provide BS-seq results in bedmethyl format using --bs_bedmethyl:
nextflow run ~/tools/longmethyl -profile singularity \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gz \
--eval_methcall true \
--bs_bedmethyl hg002_bsseq_chr20_demo.bedExtra 2. Resume a run
Try -resume to re-run a failed job to save time:
nextflow run ~/tools/longmethyl -profile singularity \
--dsname test \
--genome chr20_demo.fa \
--input fast5_chr20.tar.gz \
-resumeOutputs
The output directory should look like the following:
longmethyl_results/
├── pipeline_info
│ ├── execution_report_2022-11-12_10-33-35.html
│ ├── execution_timeline_2022-11-12_10-33-35.html
│ ├── execution_trace_2022-11-12_10-33-35.txt
│ └── pipeline_dag_2022-11-12_10-33-35.svg
└── test-ds
├── test_deepsignal_eval_genomelevel.forplot.txt
├── test_deepsignal_eval_genomelevel.txt
├── test_deepsignal_eval_readlevel.txt
├── test_deepsignal_per_read_combine.tsv.gz
└── test_deepsignal_sitemods_freq.bed.gz
- pipeline_info: Information of the workflow execution, generated by nextflow automatically.
- test-ds: methylation calling results
- test_deepsignal_eval*: Read-level/genome-level evaluation results when
--eval_methcalland--bs_bedmethylis set. - test_deepsignal_per_read_combine.tsv.gz: Per-read methylation prediction
- test_deepsignal_sitemods_freq.bed.gz: Genome-level methylation frequencies.
- test_deepsignal_eval*: Read-level/genome-level evaluation results when
Acknowledgements
developement: nextflow_develop.md
TODO
- add summary
test case with no basecall/resquiggle steps--fast5outnot necessary in basecall; tombo-anno split from tombo-resquiggle, and make it optionaldockerfilecpu settings (do not use task.cpus for all process)- clean work dir
test with gpu (with docker, run with gpu and cpu cannot succeed in a single container, cause of guppy)how to set a default deepsignal modelresult_summary_statistics/for visualization?add test demo, including benchmark and evaluationtest a 20x hg002 dataset- add deepsignal2
- add multi_to_single step
vbz issueupdate deepsignal?try filelist/multi_inputs, modify code to enable running in parallel; learn more; how to enable parallel and aviod copying files many times at the same timeDoes nextflow support cross-processes parallel (when processes have relationships in a DAG: like untar->basecall)? (maybe no)- add visualization (Rmarkdown/html?)
- freq.bed to bedgraph/wig for visualization?