 Department of Biomedical Informatics, Emory University (often stylized "Emory BMI" for GSoC communications) is committed to open source development of several biomedical informatics research projects. As a research organization, its source code lives across several open source project repositories, released with open-source licenses including BSD 3-Clause License and MIT license. Most of them can be accessed from the GitHub repositories of the research labs of Emory University School of Medicine: https://github.com/Emory-HITI, https://github.com/sharmalab, https://github.com/NISYSLAB, and https://github.com/controlcore-project/
Department of Biomedical Informatics, Emory University (often stylized "Emory BMI" for GSoC communications) is committed to open source development of several biomedical informatics research projects. As a research organization, its source code lives across several open source project repositories, released with open-source licenses including BSD 3-Clause License and MIT license. Most of them can be accessed from the GitHub repositories of the research labs of Emory University School of Medicine: https://github.com/Emory-HITI, https://github.com/sharmalab, https://github.com/NISYSLAB, and https://github.com/controlcore-project/
Emory BMI has been a successful mentoring orgnaization for Google Summer of Code 2012 - 2016, 2019, 2021, and 2022! In the recent years, we had 8 great contributors in 2022 and 6 in 2021. We are excited and looking forward to working with another batch of contributors for the upcoming GSoC. Emory BMI takes pride in having the past successes and GSoC contributors turning into long-term collaborators and mentors themselves. Emory BMI also encourages the contributors to collaborate further towards research outcomes, alongside their coding, as most of our GSoC projects include a fair amount of research.
We have been using Slack as the primary medium of communication. Since Slack has limited the features of the Slack free versison, we are slowly moving away from Slack and asking the contributors to communicate via the discussion forums of each project and this central repository instead. These discussion forums also give the contributors the potential to reach a larger audience through their public discussions, therefore, providing more transparency to our open source development discussions.
Please refer to the contributor guidelines for more details on how to apply and a standard template for the application. The ideas list is given below.
Discuss the project on the project's discussion forum (as listed below under each project idea), and once you are ready to submit your application, use the template below. You must submit your application directly using the GSoC Program Site. If you have a project idea that is relevant for Emory Biomedical Informatics, but is not listed here, feel free to consult the mentors to discuss your own idea. The ideas listed below can be open for interpretation. Feel free to discuss with the mentors for clarifications, questions, or alternative suggestions.
The ideas are marked easy, medium, and hard in difficulty level. They are also tagged 175 hours, 350 hours, or 350 hours / 175 hours. These three values represent the medium-sized projects, large projects, and the projects that have the flexibility to be adopted as a medium-size project or extended as a large project.
[1] An Integrated CONTROL-CORE experience.
Mentors: Nan Li (nan.li -at- emory.edu), Mayuresh Kothare (mvk -at- lehigh.edu), and Mark Arnold (markgarnold -at- yahoo.com)
Overview: concore is a lightweight framework for closed-Loop peripheral neuromodulation control systems. concore-editor is a React-based front-end environment for concore. concore-editor is a fork of DHGWorkflow. DHWorkflow was developed as a visual directed hypergraph editor for workflows with loops in GSoC 2021. A GSoC 2022 project forked this repository to adapt it as a front-end for concore. concore has a server back-end developed with Python Flask, which should be invoked by concore-editor as the front-end, using an approach such as the one in #59 as the first step.
concore and concore-editor are part of the CONTROL-CORE project. This project should not be limited to #59. Rather this project should provide a unified experience for control systems developers with the CONTROL-CORE project.
This GSoC aims to bring a unified experience for CONTROL-CORE users, accessing the environment through concore-editor, to create and run the studies visually.
Current Status: Although we currently have the concore-editor implementation which aims to be the front-end for concore, it has not been integrated with concore. Currently, the "Save" button allows saving a currently opened concore study back to its original location in the local filesystem where concore resides, when we use Chrome or Edge as the browser.
Currently, there is an option "Server" that introduces several buttons (Build, Debug, Run, Clear, Stop, and Destroy). But these buttons don't do anything when clicked.
Expected Outcomes: We propose a similar integration (to that of the "Save" button in concore-editor) for all the concore methods (such as Build, Debug, Run, Clear, Stop, and Destroy). That will eliminate the need to use the commandline to invoke these methods, thus providing a browser-based integrated experience through concore-editor.
A complete integrated front-end for concore, with a seamless user experience through the visual concore-editor. By the end of the GSoC, we also expect the currently identified issues to be resolved, along with any bugs that will be uncovered along the duration of the project.
Please pay attention to testing and documentation in the proposal and include sufficient time for both in the proposed timeline. As this project will provide an enhanced user experience, it is important that such improvements and changes are documented adequately. The developments should be tested across the supported operating systems and browsers to document requirements, shortcomings, and pending issues adequately.
Required Skills: Javascript (including React) and Python. This is a front-end development project, and the contributors are expected to demonstrate their previous experience and potential to implement a front-end system with Javascript frameworks such as React.
Code Challenge: Pull requests to fix concore-editor bug reports are encouraged. Demonstration of local installation of concore editor through screenshots in the proposal is expected. Please host your own forked version of concore-editor, following the "Deploying concore-editor in GitHub" section in the concore-editor README. Please also make sure to keep your hosted concore-editor instance updated with your own changes. That would allow us to confirm that your changes are working even before reviewing/testing your commits and merging your fixes to the concore-editor repository. Please also include the link of your hosted concore-editor in your proposal. By default, it will take the form of https://YOUR-GIT-REPO.github.io/concore-editor/.
Source Code: https://github.com/ControlCore-Project/concore-editor and https://github.com/ControlCore-Project/concore
Discussion Forum: https://github.com/ControlCore-Project/concore-editor/discussions
Effort: 350 Hours
Difficulty Level: Medium
[2] A real-time computing support for concore.
Mentors: Mark Arnold (markgarnold -at- yahoo.com), Mayuresh Kothare (mvk -at- lehigh.edu), and Parisa Sarikhani (psarikh -at- emory.edu)
Overview: concore is a lightweight framework for closed-Loop peripheral neuromodulation control systems. concore supports composing control systems studies from programs implemented in Python, Matlab, Verilog, and C++.
Current Status: The current implementation of concore works well for the neuromodulation control systems simulations. However, the current implementation is not optimized for real-time execution of time-sensitive executions such as real-time live experiments with organ systems, as the latency must be much lower with higher performance requirements. There are three obstacles to a full real-time implementation: i) the language chosen must support real-time computations. There is a preliminary C++ implementation here. ii) currently concore uses file sharing to communicate between processes, which may impact the real-time guarantees. iii) the software needs to run on some kind of hardware platform that supports real-time computations.
Expected Outcomes: We propose an implementation intended for such real-time executions, deployed on a real-time operating system. This implementation should provide a significant performance enhancement compared to the current implementations.
A cheap option to overcome the hardware platform problem, which could be available to many potential contributors, is the Raspberry Pi. Although not ideal for real-time executions, it appears some have used it successfully [1, 2, 3]. The contributors are encouraged to elaborate on their choice of implementation and evaluation in their proposal.
Required Skills: C++, Linux/POSIX for real-time computations, Raspberry Pi, and Python
Code Challenge: Prior experience in Python and C++ must be demonstrated. Also, a patched real-time Linux platform, which has concore running on it, must be demonstrated. Running concore on Raspberry Pi could be a small test for someone who has access to a Rapberry Pi.
Source Code: https://github.com/ControlCore-Project/concore
Discussion Forum: https://github.com/ControlCore-Project/concore/discussions
Effort: 350 Hours
Difficulty Level: Hard
[3] Making ZeroMQ a first-class feature of concore.
Mentors: Mark Arnold (markgarnold -at- yahoo.com) and Pradeeban Kathiravelu (pradeeban.kathiravelu -at- emory.edu)
Overview: concore is a lightweight framework for closed-loop peripheral neuromodulation control systems. concore consists of its own file-sharing based concore protocol to communicate between the programs in a study. We have introduced an osparc-control based communication as an alternative to this default file-sharing based concore protocol.
osparc-control is an extension of ZeroMQ. osparc-control based implementation replaces the file-sharing mechanism restricted to one local machine with message queues that can be transmitted between locally networked machine. This osparc-control based communication should be promoted as a first-class approach to implement the edges of concore.
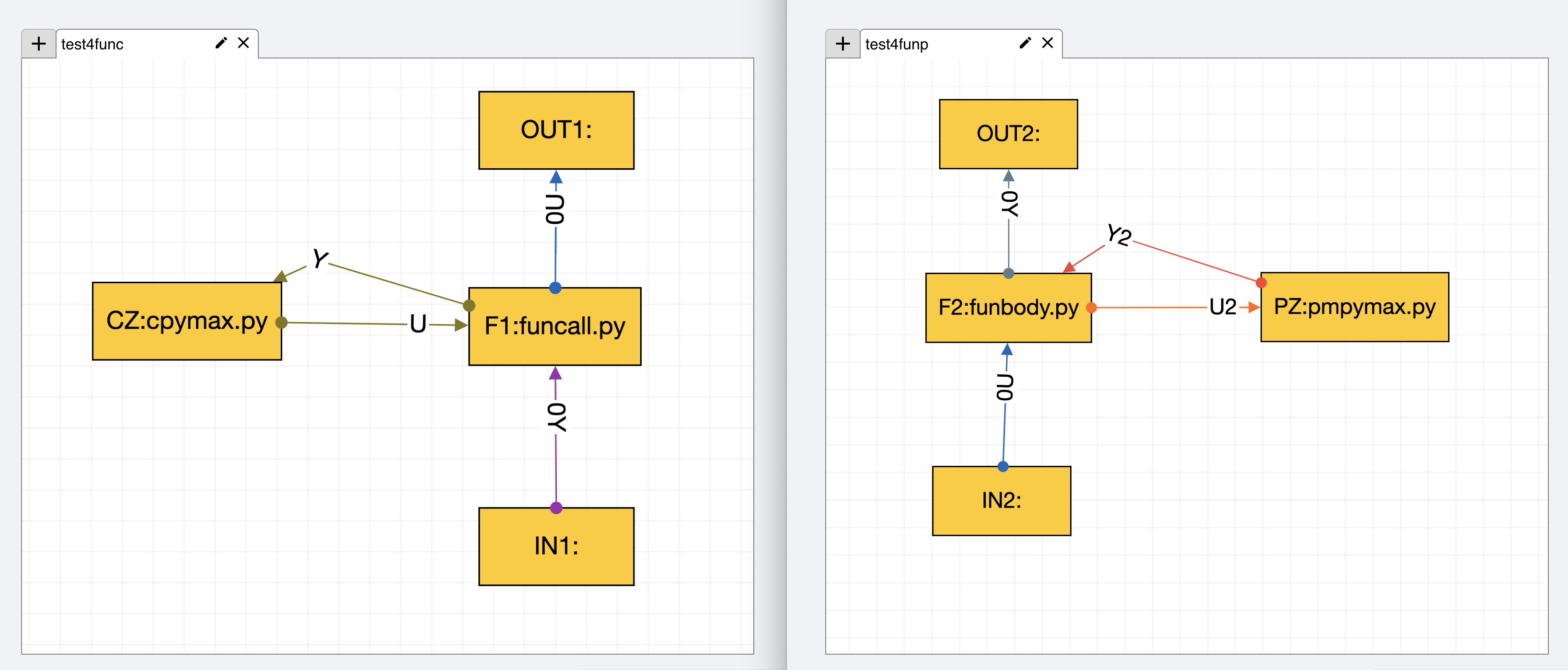 Current Status: Currently, a simple osparc-control based implementation exists in concore. However, these ZeroMQ edges are not visible in concore editor, the browser-based visual editor for concore. Consequently, studies with ZeroMQ edges are represented as forests instead of directed hypergraphs, due to the "invisible" ZeroMQ edges. This also means, to run a concore study with ZeroMQ-based edges, we have to run each hypergraph in the forest separately.
Current Status: Currently, a simple osparc-control based implementation exists in concore. However, these ZeroMQ edges are not visible in concore editor, the browser-based visual editor for concore. Consequently, studies with ZeroMQ edges are represented as forests instead of directed hypergraphs, due to the "invisible" ZeroMQ edges. This also means, to run a concore study with ZeroMQ-based edges, we have to run each hypergraph in the forest separately.
Expected Outcomes: We need to promote a unified experience in concore, whether the edges are actually implemented via the default file-sharing approach, or through this experimental osparc-control/ZeroMQ message-based approach. To illustrate the ZeroMQ-based edges, the contributor can choose to introduce a new assumption that all the ZeroMQ-edges must start with "0" in their labels. Usually, we label the edges with alphabetical characters. Therefore, this is a safe assumption. Once such a graph with ZeroMQ-edges is made (a single directed hypergraph, rather than a forest with disjoint two or more directed hypergraphs), we should be able to seamlessly build and run the study regardless of the underlying communication mechanism. Thus, we aim to demonstrate the possibility of a seamless local vs. distributed execution in a cluster through ZeroMQ.
Required Skills: Python
Code Challenge: Prior experience in Python must be demonstrated. Prior experience with message-oriented middleware frameworks such as ZeroMQ can be a plus, although not mandatory.
Source Code: https://github.com/ControlCore-Project/concore
Discussion Forum: https://github.com/ControlCore-Project/concore/discussions
Effort: 350 Hours
Difficulty Level: Medium
[4] A Reference Implementation for concore Library in Java or Julia.
Mentors: Mark Arnold (markgarnold -at- yahoo.com) and Pradeeban Kathiravelu (pradeeban.kathiravelu -at- emory.edu)
Overview: concore is a lightweight framework for closed-Loop peripheral neuromodulation control systems. Currently, it supports implementations of programs in Python, C++, Matlab, Octave, and Verilog. In this project, the contributor will develop a reference implementation of the concore library in Java or Julia.
Current Status: We developed the concore library initially in Python, and then implemented support for other languages. The contributor will work towards a reference implementation in Java or Julia in this project. The successful completion of this project will expand the user base of concore to include Java/Julia developers.
Expected Outcomes: A complete reference implementation of the concore Library in Java or Julia.
Required Skills: i) Java or Julia and ii) Python
Code Challenge: Demonstration of previous expertise in Java/Julia and Python can be beneficial.
Source Code: https://github.com/ControlCore-Project/concore
Discussion Forum: https://github.com/ControlCore-Project/concore/discussions
Effort: 175 Hours
Difficulty Level: Medium
[5] Creating shareable "albums" from Niffler data sets
Mentors: Ananth Reddy (bananthreddy30 -at- gmail.com), Hari Trivedi (hari.trivedi -at- emory.edu), and Judy Gichoya (judywawira -at- emory.edu)
Overview: Niffler is a framework to retrieve DICOM images from PACS real-time as a DICOM stream as well as retrospectively. Images can be retrieved from a PACS via Niffler in real-time (via Niffler meta-extraction module) or on-demand (via Niffler cold-extraction module). However, these downloaded data sets remain in the local environments such as a research server or a cluster where Niffler is run from. To use this data, researchers must identify certain subsets of data. This can be achieved by querying the retrieved data. For instance, Niffler stores the metadata of the data retrieved in real-time in a Mongo database. By querying the metadata, subsets of images can be identified. However, currently Niffler does not possess the ability to create such "albums" from a set of DICOM images retrieved by Niffler, and share with other users.
Current Status: Currently, Niffler does not have the ability to select subsets of images or create albums. We are sharing images through other orthogonal approaches (via rclone, for example).
Expected Outcomes: There are several approaches to implement such albums feature. One approach is to using Kheops to provide an interface to create and view the albums. MEDIator can be extended and incorporated to Niffler to create subsets and share the images via a unique URL as well.
The proposed feature will make the images retrieved by Niffler accessible by more researchers for their experiments, by replacing the current manual efforts of data sharing. Moreover, Kheops natively integrate with OHIF Viewer. As such, images retrived by Niffler can be viewed through OHIF Viewer, by creating albums with Kheops.
An approach to creating shareable datasets from the DICOM images retrieved by Niffler. It could be adopting existing frameworks such as MEDIator and Kheops and scripts and integration code with those frameworks or an entirely new module to Niffler for this feature. However, contributors are encouraged to use Kheops or alternatives, rather than reinventing the wheel (unless there is a convincing reason).
Required Skills: Python and Java.
Code Challenge: A demonstration of potential integration of Niffler with such existing frameworks. The proposed frameworks are samples only. The contributors may choose their own.
Source Code: https://github.com/Emory-HITI/Niffler/
Discussion Forum: https://github.com/Emory-HITI/Niffler/discussions
Effort: 175 Hours
Difficulty Level: Easy
[6] Develop a drag-and-drop frontend for WDL and CWL workflows
Mentors: Babak Mahmoudi (b.mahmoudi -at- emory.edu) and Özgür Kara (ozgurrkara99 -at- gmail.com).
Overview: Standard workflow languages such as Common Workflow Language (CWL) and Workflow Description Language (WDL) are traditionally written by hand and executed by workflow frameworks such as Cromwell and Toil. Drag-and-drop front-end frameworks exist, but are also limited limited by their usability across platforms. A seamless front-end to support workflow development can help the open source community tremendously. CWL and WDL limit their focus to workflows that can be represented by a directed acyclic graph (DAG). So, while a drag-and-drop interface may allow more diverse graph types such as directed graphs, when converting, the DAG format must be verified.
Current Status: Our previous GSoC project DHGWorkflow enabled us to visually create Directed Hypergraphs (DHGs) and export them as GraphML files through its browser-based lightweight environment. This project could use a similar approach to generate WDL and CWL files from a browser-based application. Forking and adopting DHGWorkflow is an option. There are also stand-alone CWL and WDL drag-and-drop projects such as Rabix that can be adopted for this project.
Expected Outcomes: APIs (such as RESTful interfaces) should be provided to internally pass the workflow definitions to the backend, to avoid having a backend application having to read and parse the workflow files (WDL and CWL) again, rather than having them directly use the workflow definitions by the other programs.
Required Skills: Languages of choice for front-end and the APIs.
Code Challenge: A demo to highlight the potential of developing this task, such as a prototype or a mock up.
Source Code: New Project
Discussion Forum: https://github.com/NISYSLAB/Emory-BMI-GSoC/discussions
Effort: 350 Hours
Difficulty Level: Medium
[7] A Framework for Unsupervised Deep Clustering
Mentor: Mahmoud Zeydabadinezhad (mzeydab -at- emory.edu) and Babak Mahmoudi (b.mahmoudi -at- emory.edu)
Overview: Clustering is a fundamental task in machine learning and data mining, with a wide range of applications such as image and speech recognition, anomaly detection, and natural language processing. Traditional clustering methods, such as k-means and hierarchical clustering, are based on shallow models and rely on hand-engineered features. In recent years, deep learning techniques have been applied to clustering, resulting in improved performance and the ability to automatically learn features from the data. However, existing methods are mostly supervised and require labeled data, which is not always available. Additionally, they are often not designed to handle low-resource scenarios.
The objective of this project is to develop an open-source framework that utilizes unsupervised deep learning techniques for data clustering. The framework should be capable of handling low-resource scenarios and be able to scale to large datasets.
Current Status: New project.
Expected Outcomes: The proposed framework will utilize unsupervised deep learning techniques for data clustering. Specifically, the framework will be based on autoencoder networks, which can be trained to learn a compact, low-dimensional representation of the data. The encoded data will then be used as input to a clustering algorithm. To handle low-resource scenarios, the framework will also incorporate techniques such as active learning and transfer learning. The framework will be implemented in an open-source programming language, such as Python, and will be made publicly available on a platform such as GitHub.
Data: The framework will be evaluated on a publicly available electroencephalograpghy (EEG) data. Evaluation: The performance of the proposed framework will be evaluated using metrics such as normalized mutual information. The framework will also be compared to existing state-of-the-art methods for unsupervised deep clustering. Outline:
Literature Review: Research and review existing unsupervised deep clustering methods for medical data, specifically EEG data. Identify the current limitations and challenges in this field.
Data Preprocessing: Develop preprocessing techniques to prepare EEG data for unsupervised deep clustering. This includes filtering, denoising, and feature extraction.
Clustering Framework: Design and develop an unsupervised deep clustering framework that can handle medical data, specifically EEG data. This framework should be able to extract meaningful patterns and insights from the data in an unsupervised manner.
Evaluation: Evaluate the performance of the developed framework using a variety of metrics and benchmarks. Compare the results to existing unsupervised deep clustering methods for medical data.
Deployment: Create an open-source implementation of the developed framework and make it available for the community to use. Applications: Explore the potential applications of the developed framework on various medical data, specifically EEG data. This includes, but is not limited to, epilepsy diagnosis, brain-computer interface and sleep stage classification.
Required Skills: Python and deep learning
Source Code: New Project
Discussion Forum: https://github.com/NISYSLAB/Emory-BMI-GSoC/discussions
Effort: 350 Hours
Difficulty Level: Hard
[8] Visualizing MRI scanner utilization on-demand from DICOM metadata
Mentors: Nan Li (nan.li -at- emory.edu), Puneet Sharma (puneet.sharma -at- emory.edu), and Ryan Birmingham (rbirmin -at- emory.edu)
Overview: Niffler enables computing scanner utilization using DICOM metadata, from DICOM images received on-demand or in real-time. DICOM tags are used to compute scanner utilization on-demand by the SUVPaR module.
The typical execution workflow for the on-demand execution is: cold-extraction (retrieve images on-demand from the PACS to the research cluster) -> png-extraction (extract all the DICOM attributes from the images in a CSV file) -> SUVPaR (compute scanner utilization from the csv file and produce an output CSV file).
Current Status: While we have a scanner utiliization computed in the backend, there is no integrated front-end to present the results elegantly. The created results are currently stored in CSV files and displayed through an Eaglescope dashboard.
There is a Slurm-based experimental distributed implementation for the png-extraction in ImageExtractorSlurm.py. This implementation aims to make png-extraction to work on a cluster, rather than a single server. However, this implementation is outdated as it did not get updated with the latest developments that happened on png-extraction.
Expected Outcomes: The series-level scanner usage computations can be resource-heavy, as it requires DICOM metadata processing of a lot of images (more than computing at just the study-leve) and then processing the metadata to compute scanner usage. This project should optimize the executions of png-extraction and suvpar as needed towards this goal.
Potential to use a cluster, instead of a single server instance for the metadata extraction can be considered. The slurm-based png-extraction may be useful in a distributed execution of png-extraction, and similar approaches should be considered for speedup of the execution. Some practical approaches to performance enhancement could be, optimizing the queries (at cold-extraction and/or png-extraction) and filtering based on scanners early on (for example, at cold-extraction and/or png-extraction), to minimize the problem size early on.
The front-end should be improved with a better integration to visualize the scanner usage at series level. The existing Eaglescope-based dashboard can be used as a starting point for this. The output CSV file from SUVPaR should be adopted to be consumed by the Eaglescope-based dashboards for the visualization.
Finally, scanner usage metrics at series level should be seamlessly presented via an integrated dashboard by end of this GSoC.
Required Skills: Python and Javascript. Prior experience with DICOM would be a plus.
Code Challenge: A bug fix from Niffler can be a positive indication of understanding the code base.
Source Code: https://github.com/Emory-HITI/Niffler/
Discussion Forum: https://github.com/Emory-HITI/Niffler/discussions
Effort: 350 Hours
Difficulty Level: Hard
[9] Auto-detect coverage bounding boxes for brain MRI images
Mentors: Puneet Sharma (puneet.sharma -at- emory.edu) and Tony Pan (tony.pan -at- emory.edu)
Overview: In this project, the contributor will develop a methodology to auto-detect coverage bounding boxes for the brain and other body parts using image processing. For example, detect the top of the head to the lowest extent (~C2/3) and left-to-right extents. This module will then be used to assess coverage compliance on MRI modalities.
Current Status: This is a new module to run on DICOM images, specifically brain MRI images in DICOM format. This module will execute on DICOM images acquired in real-time or on-demand by Niffler. For testing purposes during the application period and early stages of development, brain MRI images obtained from public data sources such as the Cancer Imaging Archive (TCIA) can be used.
Expected Outcomes: This project builds an image processing methodology to autodetect coverage bounding boxes on brain MRI images. The algorithm will be trained on the brain (or other body parts) data and tested on more data of the same body parts for validation for accuracy.
Required Skills: Python. Prior experience with DICOM would be a plus.
Code Challenge: Experience working with machine learning libraries and similar problems is expected. Candidates are encouraged to include links/pointers to code samples or similar projects to highlight their experience in their proposal.
Source Code: https://github.com/Emory-HITI/Niffler/
Discussion Forum: https://github.com/Emory-HITI/Niffler/discussions
Effort: 350 Hours
Difficulty Level: Hard
[10] A "contribute" action to concore studies.
Mentors: Pradeeban Kathiravelu (pradeeban.kathiravelu -at- emory.edu), Mark Arnold (markgarnold -at- yahoo.com), Tony Pan (tony.pan -at- emory.edu), and Babak Mahmoudi (b.mahmoudi -at- emory.edu).
Overview: concore is a lightweight framework for closed-Loop peripheral neuromodulation control systems. concore supports composing control systems studies from programs implemented in Python, Matlab, Verilog, and C++. concore also consists of a set of programs and studies that are already developed by the contributors (can be seen from the demo, testsou, ratc, and humanc folders in the concore repository). We envision more community-contributed studies and programs in the future.
Current Status: Currently, to contribute a study or a program to concore, a user must fork the concore repository and send a pull request through Git. This contribute approach does not help with users that are not familiar with Git or GitHub. The concore-editor has a "Contribute" button, which does nothing at this point.
Expected Outcomes: Once we have this project successfully completed, concore will have a script named "contribute" with necessary GitHub actions (or a similar alternative approach). With this Contribute option, the contributors merely invoke the contribute script and a pull request is generated or updated on their behalf. This project also has a research component, and the contributors are encouraged to propose their alternative approach (if any) in their proposal.
The contribute script will take user inputs user ID (likely, GitHub ID, to assign with the commit logs) and study name/identifier, in addition to the location of the study and programs to be contributed. Therefore, each user can contribute their studies with a study name that is locally unique. That means, while there could be multiple "human_cardiac_model," a pair of {pradeeban, human_cardiac_model} will be unique. First mention of this pair as input to the contribute script will create a pull request as a new study. When such a study already exists, subsequent executions will create an update through a pull request to an existing study that is already contributed by the same user ID. Measures should be taken to ensure that the user ID is truly unique.
The major contribution of this project is the contribute functionality to be implemented as a backend script in concore as described above. Then, the currently stub "Contribute" button in the concore-editor can be configured to invoke the local contribute script implemented as part of this GSoC. By end of the GSoC, the contributor can work on that integration between the "Contribute" button in concore-editor and the "contribute" script implemented for concore. The integration of these two should be similar to the other methods (run, debug, build, ... ) of concore-editor, which are currently not integrated into the concore backend as well.
Required Skills: Python
Code Challenge: Prior experience in Python with code contributions to highlight, with attempts at bug fixes / pull requests to concore repository.
Source Code: https://github.com/ControlCore-Project/concore
Discussion Forum: https://github.com/ControlCore-Project/concore/discussions
Effort: 350 Hours
Difficulty Level: Medium
[11] A methodology to categorize exam series.
Mentors: Tony Pan (tony.pan -at- emory.edu) and Puneet Sharma (puneet.sharma -at- emory.edu)
Overview: This project aims to develop methodology to categorize exam series from a variety of vendors/systems to a standardized protocol template. This is an important step to begin assessing protocol compliance of DICOM image series. The project focuses on radiographic images of MRI modality.
Current Status: This is a new module to run on DICOM images of MRI modality. This module will execute on DICOM images acquired in real-time or on-demand by Niffler. For testing purposes during the application period and early stages of development, MRI images obtained from public data sources such as the Cancer Imaging Archive (TCIA) can be used.
Expected Outcomes: There would be no evaluation of images or quality. The task would be to identify whether the series belonged to a defined protocol, and if so whether its DICOM parameters meet certain threshold values.
Required Skills: Python. Prior experience with DICOM would be a plus.
Code Challenge: Candidates are encouraged to include links/pointers to code samples or similar projects to highlight their experience in their proposal.
Source Code: https://github.com/Emory-HITI/Niffler/
Discussion Forum: https://github.com/Emory-HITI/Niffler/discussions
Effort: 350 Hours
Difficulty Level: Hard