Nebula is an ultra-efficient mapping-free structural variation genotyper based on kmer counting.
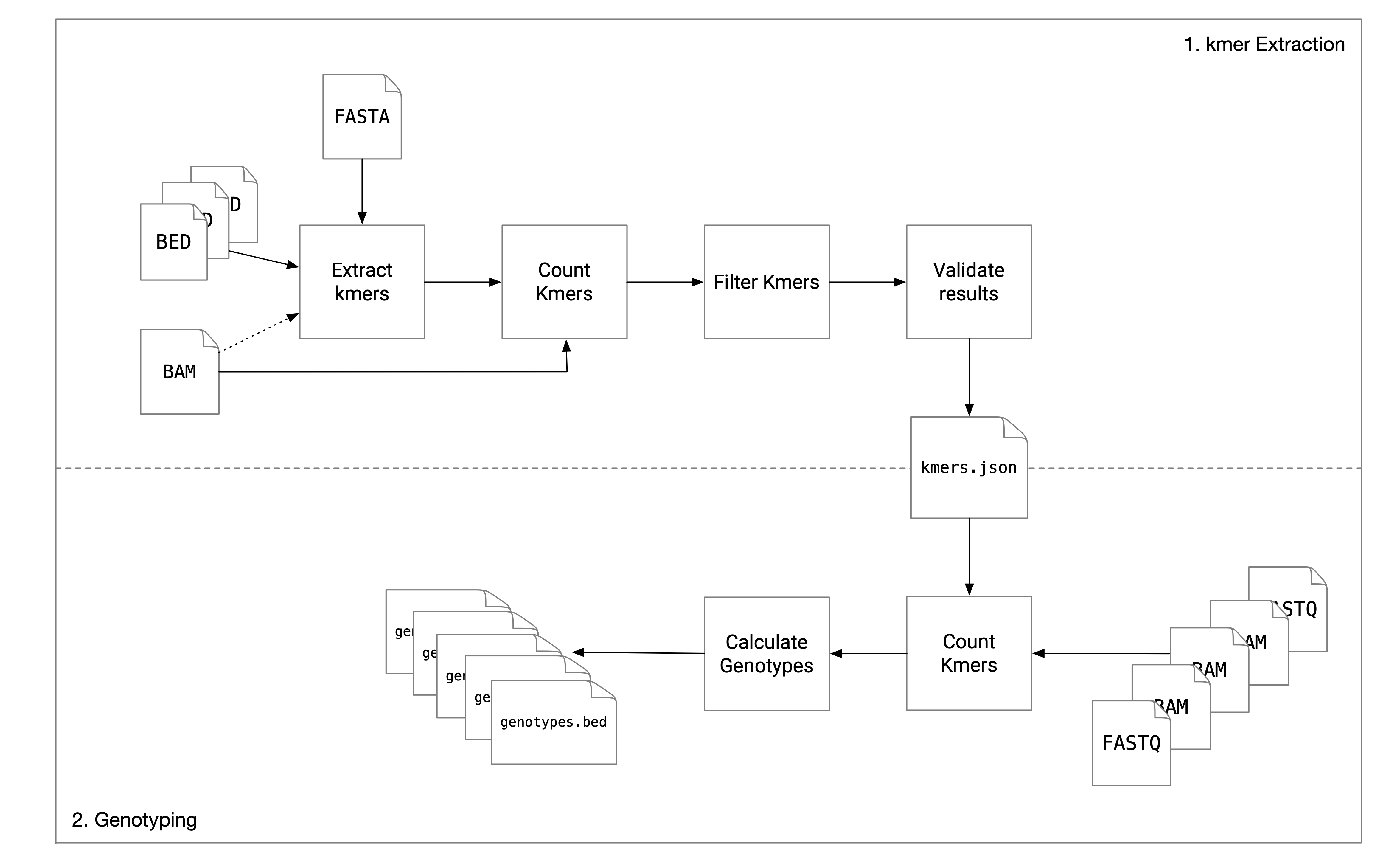
Nebula is a mapping-free approach for accurate and efficient genotyping of SVs. Nebula is a two-stage approach and consists of a kmer extraction phase and a genotyping phase. Given as input a set of SV coordinates (BED/VCF), the reference assembly (FASTA), and a set of mapped samples on which the genotype of these SVs is already known, Nebula extracts a collection of kmers that represent the input SVs (kmer extraction phase) and these extracted kmers will then be used to genotype the same set of SVs on any new WGS sample(s) without the need to map the reads to the reference genome (genotyping phase). This is done by counting the kmers in the WGS reads of the new sample(s) and predicting genotypes using a likelihood model.
Nebula's only dependency is htslib which is included as submodule for compatibility reasons. Clone recursively:
git clone --recursive git@github.com:Parsoa/Nebula.git
cd Nebula
Then build htslib:
cd src/cpp/htslib
make
Then build Nebula itself:
cd ..
make
You need to update your LD_LIBRARY_PATH to use Nebula's own htslib instance:
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/path/to/Nebula/src/cpp/htslib
Nebula receives the SV loci and genotypes for prepreocessing in BED format. Only a few fields are required:
#CHROM BEGIN END SVTYPE SEQ GENOTYPE
The header line MUST be present.
SVTYPE is usually one of DEL, INS or INV for deletion, insertion and inversion respectively. Other SV types are not currently tested but should work as long as kmers can be extracted for them.
The SEQ field is only required for insertions and is ignored for insertions and inversions, however the field should still be provided (you can pass an empty string ".").
You can optionally include SVLEN otherwise it will be estimated from BEGIN and END for deletions and inversions and from size of SEQ for insertions.
Chromosome names in BED and FASTA files can both include and not include chr. However, Nebula's output will always include chr in chromosome names regradless of input format.
GENOTYPE must be one of 0/0 for absent, 0/1 or 1/0 (treated equally) for heterozygous and 1/1 for homozygous. Nebula's preprocessing stage requires "known" SV genotypes to select kmers, as a result ambiguous genotypes like ./1 and ./. are not accepted and will result in abort.
Nebula requires a reference genome in FASTA format and a number of samples in BAM/SAM/CRAM format along with SV genotypes for each sample in BED format. k-mers are extracted from both the reference genome (for deletions only) and read mappings (for any type of SV).
The same set of SV genotypes should be provided for all samples being preprocessed. If the set of SVs is not the same, then the union of all SVs found in the BED files will be considered and SVs that don't have a genotype for a sample are assumed to be 0/0 on that sample.
The preprocessing stage consists of multiple commands. To extract kmers run the preprocess command. This will output the extracted kmers in a series of JSON files under --workdir and the directory is created if it doesn't exists. A BED file and a BAM file should be passed for each sample being preprocessed.
nebula preprocess --bed /path/to/genotypes_1.bed,/path/to/genotypes_2.bed --bam /path/to/bam_file_1.bed /path/to/bam_file_2.bed --wokdir /output --reference /path/to/reference/FASTA/file --thread <number of threads to use>
Next, the input samples are genotyped with these kmers to filter low-quality kmers. Passing --select causes the genotyper to only keep the kmers whose counts support the known genotypes. --kmers specifices the location of extracted kmers (the output of running the preprocessor). The gc_kmers and depth_kmers arguments take a precaculated list of kmers used for estimating sequencing depth across regions of the genome with different levels of GC content. These files can be downloaded from here.
Passing --unique causes the genotyper to only keep kmers unique to one SV (e.g kmers that don't appear as a result of two or more different SVs). This option will usually result in higher precision when genotyping new samples but it may reduce recall slightly.
nebula genotype --bed /path_to_genotypes_1.bed --bam /path/to/bam_file_1.bed --workdir output/sample_1 --kmers /output --depth_kmers depth_kmers.json --gc_kmers gc_kmers.json --select --unique
nebula genotype --bed /path_to_genotypes_2.bed --bam /path/to/bam_file_2.bed --workdir output/sample_2 --kmers /output --depth_kmers depth_kmers.json --gc_kmers gc_kmers.json --select --unique
Finally, merge the remaining kmers after filtering by running the mix subcommand. The --samples argument takes the name of input sample. This is basically the name of the directories passed during the previous stage when running the genotyper.
nebula mix --bed /path_to_genotypes_1.bed,/path_to_genotypes_2.bed --samples sample_1,sample_2 --workdir /output
The output kmers are stored in a folder named Mix inside workdir (here /output/Mix). You will only the contents of Mix for genotyping other samples. Any other files and directories created by Nebula can be deleted to save disk space.
For genotyping unmapped sample with the extracted kmers from an earlier kmer-extraction run:
nebula genotype --kmers /path/to/Mix/directory --bam/--fastq /path/to/sample --workdir <output directory>
Note that a BED file is not passed to the genotyper as the variants are implicit in the kmers.
The workdir here can be anywhere. This will count the kmers on the new sample and calculate genotypes. Note that we don't pass --select when actually genotyping a sample.
Nebula will output a BED file named genotypes.bed in the specified working directory. The file will include the original fields in the input BED files along with the field GENOTYPE (one of 0/0, 1/0 or 1/1).
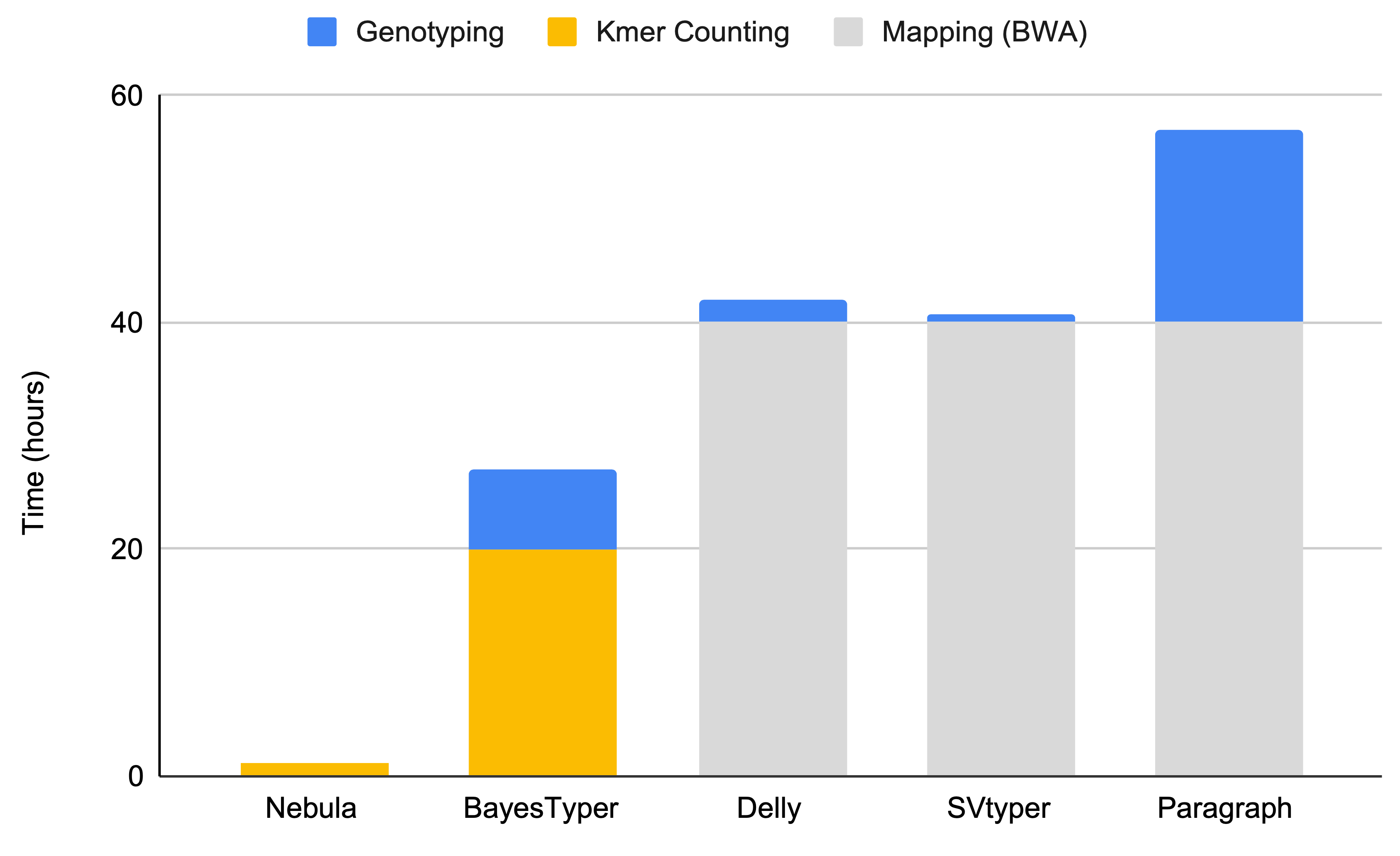
Nebula is designed to be simple, fast and memory efficient so it can be run on any reasonable personal hardware. Using a single processor core, Nebula can count kmers at a rate of 400,000 reads per second from a FASTQ file. A 30x human sample can be process in less than 80 minutes on a single core.
Nebula has been deeply parallelized with OpenMP. The kmer extraction step (preprocess subcommand) will benefit significantly from multiple threads as finding occurrences of kmers in sequencing reads and the reference genome can be easily parallelized. Use as many threads as available for the preprocess subcommand for best performance.
Nebula's kmer counter is very fast due to low-level optimizations and as a result genotyping runtime is mostly a function of memory bandwidth and I/O speed. Unless you have very fast disk I/O, it's unlikely that adding more threads will improve kmer counting runtime. The initial loading of kmers and the final step of genotyping SVs using the likelihood model will benefit significantly from multiple threads, however they account for only 15% of the runtime. We suggest running the counter on a small number of threads (e.g 4).
Nebula is an order of magnitude faster than other tools when genotyping an unmapped sample in FASTQ sample. The figure below shows comparison of Nebula's single-core genotyping runtime against several other state-of-the-art methods.
Nebula was originally presented in Recomb-Seq 2019, the pre-print for that version can be found below:
https://doi.org/10.1101/566620
The current version of Nebula is substantially different from what was presented in 2019. Nebula was published on Oxford Nucleic Acid Research In Jan 2021: