- This repository provides code for the paper “Slimmable Transformer with Hybrid Axial-Attention for Medical Image Segmentation”
- If you have any questions about our paper, feel free to contact us.
This repository hosts the code for the following networks:
- Axial-attention (model name: attresunet)
- Axial-DeepLab (model name: axailunet)
- Gated Axial Attention U-Net (model name: gated)
- Resunet (model name: resunet )
- Unet (model name: unet )
- Slimmable Tranformer (model name: slimmable )
The transformer architecture has achieved remarkable success in medical image analysis owing to its powerful capability for capturing long-range dependencies. However, due to lack of intrinsic inductive bias in modeling visual structural information, transformer generally requires a large-scale pre-training schedule, limiting the clinical applications over expensive small-scale medical data. To this end, we propose a slimmable transformer to explore intrinsic inductive bias via position information for medical image segmentation. Specifically, we empirically investigate how different position encoding strategies affect the prediction quality of the region of interest (ROI) and observe that ROIs are sensitive to different position encoding strategies. Motivated by this, we present a novel Hybrid Axial-Attention (HAA) that can be equipped with pixel-level spatial structure and relative position information as inductive bias. Moreover, we introduce a gating mechanism to achieve efficient feature selection and further improve the representation quality over small-scale datasets. Experiments on LGG and Covid19 datasets prove the superiority of our method over the baseline and previous works. Internal workflow visualization with interpretability is conducted to validate our success better, the proposed slimmable transformer has the potential to be further developed into a visual software tool for improving computer-aided lesion diagnosis and treatment planning.
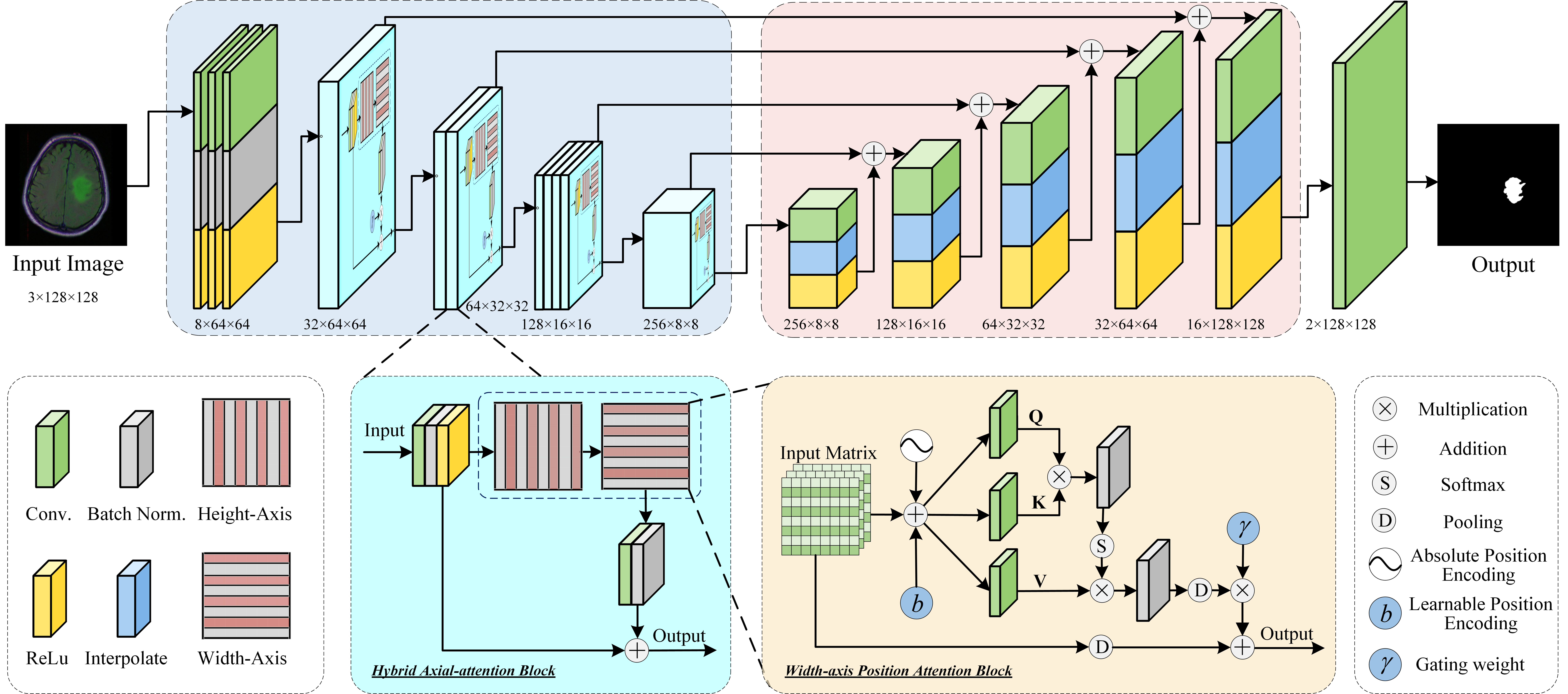
A diagram showing the architecture of the proposed model. The proposed hybrid axial-attention block is a fundamental building block of the encoder, which propagates information along the height-axis and width-axis sequentially to model the long-range dependency. A width-axis position attention block shows the entire process of calculating the attention score by combining LPE with APE.
git clone https://github.com/NanMu-SICNU/SlimmableTransformer
cd SlimmableTransformer
- Note that Slimmable Transformer is only tested on Ubuntu OS 18.04 with the following environments (CUDA-11.0). It may work on other operating systems as well but we do not guarantee that it will. To install all the dependencies using conda (The code is stable using Python 3.8, Pytorch 1.5.0) :
conda env create -f environment.yml
- To install all the dependencies using pip:
pip install -r requirements.txt
├── debug.py ├── environment.yml # environment seetings with conda └── EvaluationCode/ # performance metrics code in MATLAB └── evaluation_method/ ├── AUC_Borji.m ├── CalAUCScore.m ├── CalDice.m ├── CalDiceSenSpe.m ├── CalMeanFmeasure.m ├── CalMeanMAE.m ├── CalMeanWF.m …… ├── Testingfor_Abiation.m ├── Testingfor_Covid19.m └── lib/ # code of model and dataloader ├── build_dataloader.py ├── build_model.py ├── build_optimizer.py └── datasets/ ├── imagenet1k.py ├── metrics.py └── models/ ├── axialnet.py ├── coatten.py ├── model_codes.py ├── resnet.py ├── resunet.py ├── unet.py ├── utils.py ├── utils.py ├── metrics.py # performance metrics code in python ├── README.md ├── requirements.txt # environment seetings with pip ├── test.py ├── train.py ├── utils.py ├── utils_gray.py └── utilTools/ # common function used in data processing ├── dataAugmentation.py ├── evaluateMetric.py ├── imageTool.py ├── niito2D.py ├── weightedrfinal_model.pth
Prepare the dataset in the following format for easy use of the code. The train and test folders should contain two subfolders each: img and label. Make sure the images their corresponding segmentation masks are placed under these folders and have the same name for easy correspondance. Please change the data loaders to your need if you prefer not preparing the dataset in this format.
Train Folder-----
img----
0001.png
0002.png
.......
labelcol---
0001.png
0002.png
.......
Validation Folder-----
img----
0001.png
0002.png
.......
labelcol---
0001.png
0002.png
.......
Test Folder-----
img----
0001.png
0002.png
.......
labelcol---
0001.png
0002.png
.......
- The ground truth images should have pixels corresponding to the labels. Example: In case of binary segmentation, the pixels in the GT should be 0 or 255.
- LGG Dataset - Kaggle: Your Home for Data Science
- Covid19 Dataset - COVID-19 CT segmentation dataset
python train.py --train_dataset "enter train directory" --val_dataset "enter validation directory" --direc 'path for results to be saved' --batch_size 1 --epoch 100 --save_freq 10 --modelname "weighted" --learning_rate 0.001 --imgsize 256 --gray "no"
Change model name to unet, resunet or gated to train them (refer to About this repository), when training is completed, the weights will be saved in the path you indicate (i.e. **model_best.pth).
python test.py --loaddirec "./saved_model_path/model_name.pth" --val_dataset "test dataset directory" --direc 'path for results to be saved' --batch_size 1 --modelname "gatedaxialunet" --imgsize 256 --gray "no"
The results including predicted segmentations maps will be placed in the results folder along with the model weights. Run the performance metrics code in MATLAB for calculating 1) AUC score (Area Under Curve); 2) MAE score (Mean Absolute Error); 3) WF (Weighted F-measure) score; 4) OR (Overlapping Ratio) score; 5) Dice score; 6) Jaccard score; 7) Sensitivity; 8) Specificity; 9) Parameters (#M), 10) FLOPs (#GMac) and 11) inference time (#h).
1)Note that these experiments were conducted in RTX 3090 GPU with 128 GB memory. 2)Google Colab Code is an unofficial implementation for quick train/test. Please follow original code for proper training.
If you find our code useful for your research, please cite our paper:
Y. Hu, N. Mu, L. Liu, L. Zhang, and J. Jiang, "Slimmable Transformer with Hybrid Axial-Attention for Medical Image Segmentation", Computers in Biology and Medicine, 2024. (Under Review)
In case of any questions, please contact the corresponding author N. Mu at nanmu@sicnu.edu.cn