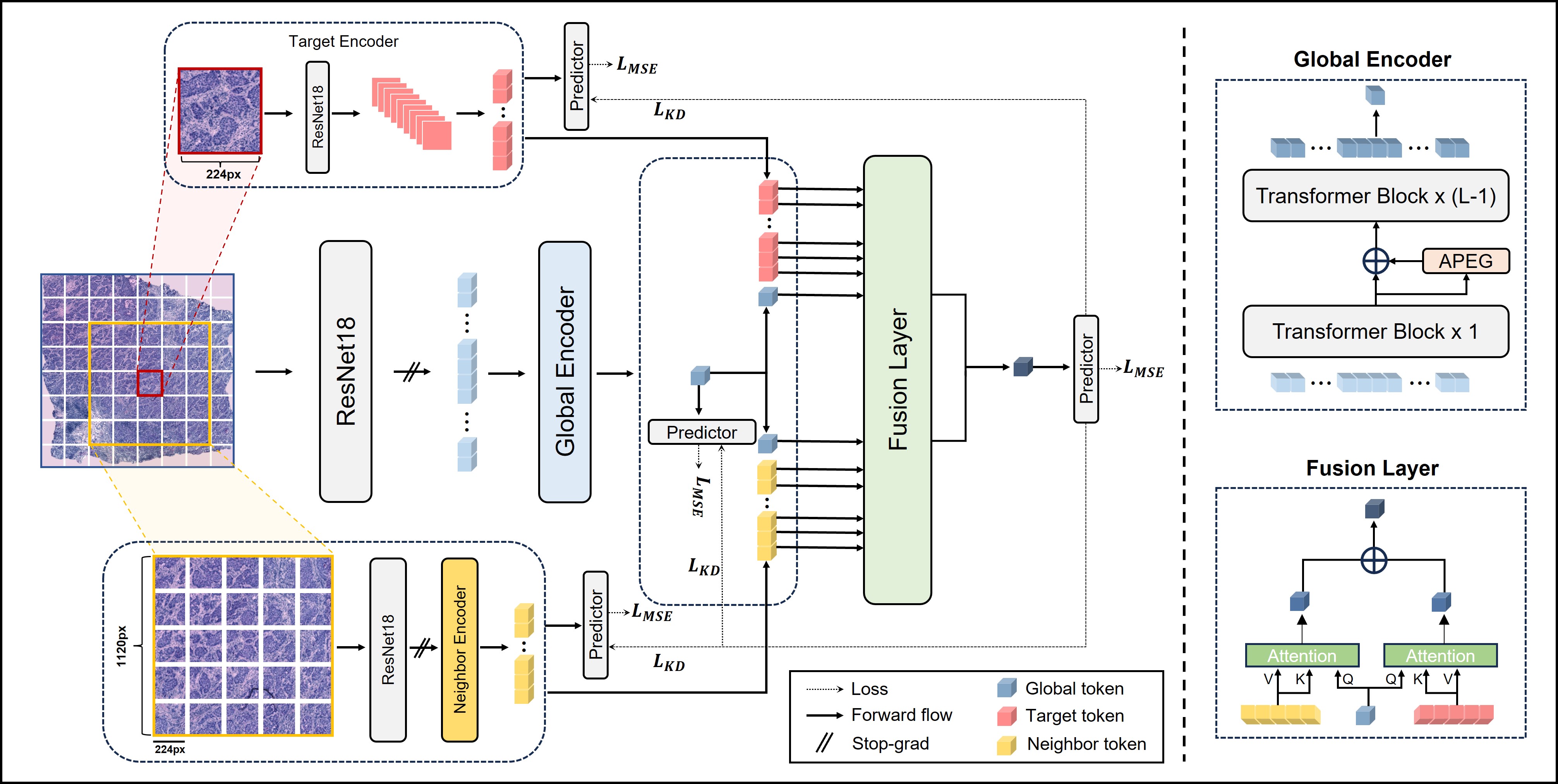
Accurate Spatial Gene Expression Prediction by integrating Multi-resolution features (accepted to CVPR 2024)
Youngmin Chung, Ji Hun Ha, Kyeong Chan Im, Joo Sang Lee*
- Add code to automatically download the ResNet18 weight
- Add code for inference
- Add code to preprocess WSIs in svs or tif format
- Python 3.9.19
- Install pytorch
pip install torch==1.13.0+cu117 torchvision==0.14.0+cu117 --extra-index-url https://download.pytorch.org/whl/cu117- Install the remaining required packages
pip install -r requirements.txtBegin by downloading the preprocessed data here.
Save the downloaded TRIPLEX.zip file into the ./data directory within your project workspace.
After downloading, unzip the TRIPLEX.zip file using the following command:
unzip ./data/TRIPLEX.zip -d ./dataThis will extract the data into four subdirectories within the ./data folder, namely her2ts, skin, stnet, and test.
TRIPLEX requires pre-extracted features from WSIs. Run following commands to extract features using pre-trained ResNet18.
- Cross validation
# BC1 dataset
python extract_features.py --config her2st/TRIPLEX --test_mode internal --extract_mode target
python extract_features.py --config her2st/TRIPLEX --test_mode internal --extract_mode neighbor
# BC2 dataset
python extract_features.py --config stnet/TRIPLEX --test_mode internal --extract_mode target
python extract_features.py --config stnet/TRIPLEX --test_mode internal --extract_mode neighbor
# SCC dataset
python extract_features.py --config skin/TRIPLEX --test_mode internal --extract_mode target
python extract_features.py --config skin/TRIPLEX --test_mode internal --extract_mode neighbor- External test
# 10x Visium-1
python extract_features.py --test_name 10x_breast_ff1 --test_mode external --extract_mode target
python extract_features.py --test_name 10x_breast_ff1 --test_mode external --extract_mode neighbor
# 10x Visium-2
python extract_features.py --test_name 10x_breast_ff2 --test_mode external --extract_mode target
python extract_features.py --test_name 10x_breast_ff2 --test_mode external --extract_mode neighbor
# 10x Visium-3
python extract_features.py --test_name 10x_breast_ff3 --test_mode external --extract_mode target
python extract_features.py --test_name 10x_breast_ff3 --test_mode external --extract_mode neighborAfter completing the above steps, your project directory should follow this structure:
# Directory structure for HER2ST
.
├── data
│ ├── her2st
│ │ ├── ST-cnts
│ │ ├── ST-imgs
│ │ ├── ST-spotfiles
│ │ ├── gt_features_224
│ │ └── n_features_5_224
└── weights/tenpercent_resnet18.ckpt
- BC1 dataset
# Train
python main.py --config her2st/TRIPLEX --mode cv
# Test
python main.py --config her2st/TRIPLEX --mode test --fold [num_fold] --model_path [path/model/weight]- BC2 dataset
# Train
python main.py --config stnet/TRIPLEX --mode cv
# Test
python main.py --config stnet/TRIPLEX --mode test --fold [num_fold] --model_path [path/model/weight]- SCC dataset
# Train
python main.py --config skin/TRIPLEX --mode cv
# Test
python main.py --config skin/TRIPLEX --mode test --fold [num_fold] --model_path [path/model/weight]Training results will be saved in ./logs
- Independent test
# 10x Visium-1
python main.py --config skin/TRIPLEX --mode external_test --test_name 10x_breast_ff1 --model_path [path/model/weight]
# 10x Visium-2
python main.py --config skin/TRIPLEX --mode external_test --test_name 10x_breast_ff2 --model_path [path/model/weight]
# 10x Visium-3
python main.py --config skin/TRIPLEX --mode external_test --test_name 10x_breast_ff3 --model_path [path/model/weight]- Code for data processing is based on HisToGene
- Code for various Transformer architectures was adapted from vit-pytorch
- Code for position encoding generator was adapted via making modifications to TransMIL
- If you found our work useful in your research, please consider citing our works(s) at:
@inproceedings{chung2024accurate,
title={Accurate Spatial Gene Expression Prediction by integrating Multi-resolution features},
author={Chung, Youngmin and Ha, Ji Hun and Im, Kyeong Chan and Lee, Joo Sang},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
pages={11591--11600},
year={2024}
}