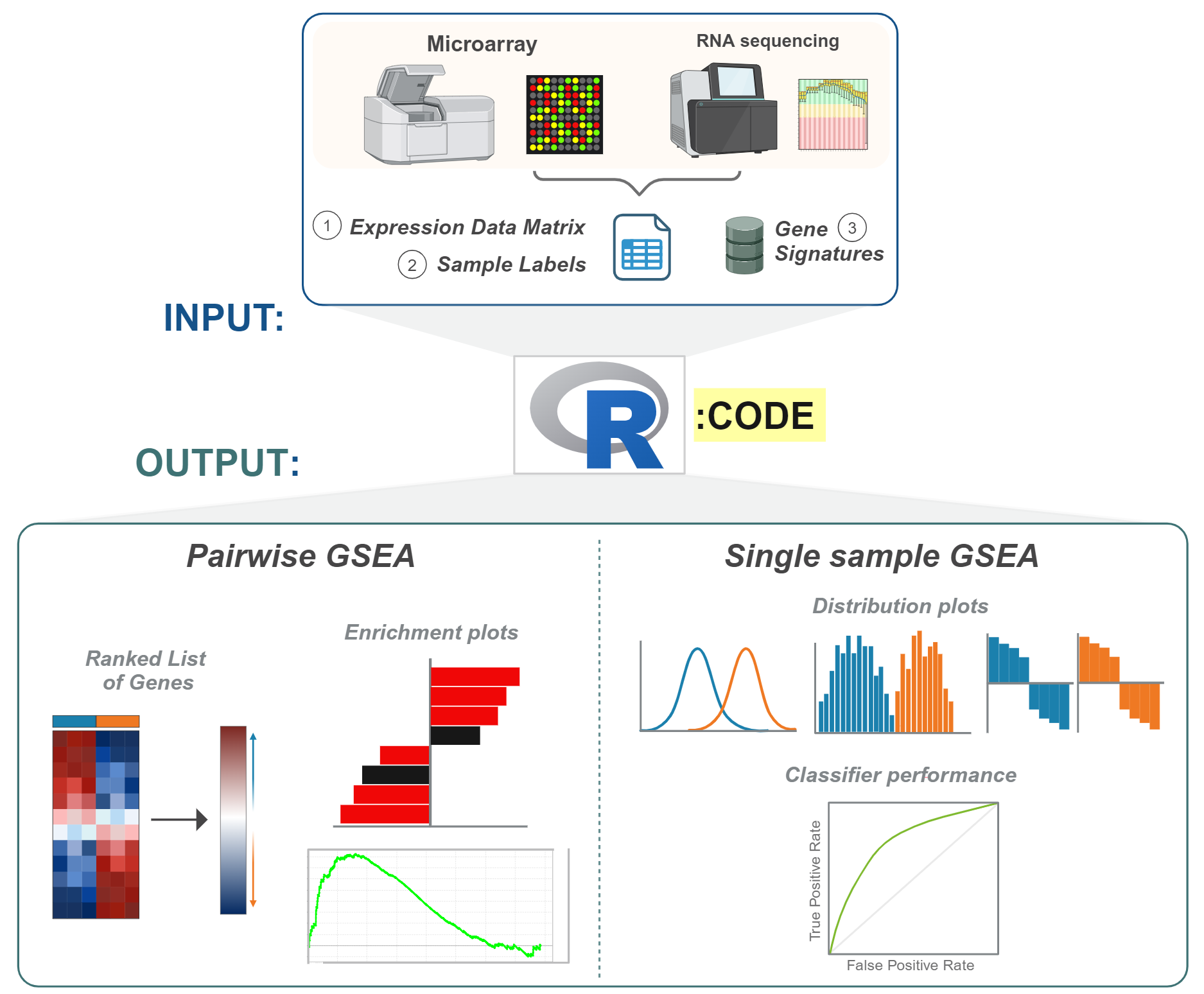
This repository consists of dualGSEA() R function created for the preprint BioRxiv submission:
"Evaluation of Gene Set Enrichment Analysis (GSEA) tools highlights the value of single sample approaches over pairwise for robust biological discovery."
Before you run the function dualGSEA(), please make sure:
- Your input data are structured accordingly:
- Expression data (Microarray or RNA-seq): As dataframe with your gene symbols as your rows and sample ids as your columns. NOTE:
dualGSEAdoes not provide option for altering parameters in the dependency functions such as forfgseaorgsvaand utilises the default parameters. NOTE: for RNA-seq data, the recommendation is you have your normalised data to run rather than raw counts (as the function relies onlimmafor ranking method and notDESeq, and currently, the function does not include the input rankings from user directly.). - Group data: Please make sure the groups are of n=2 groupings for pairwise comparisons. The data need to be so that the rownames are sample ids, so it can match up with expression data internally.
- GeneSet list: Please supply the gene signatures as a list. NOTE: Due to multiple outputs of the function, the execution and performance of the function may depend on the number of geneset list you supply. The larger the number of geneset list, the longer it may take to process.
- R packages requirement and installations: Please make sure you have all the dependencies packages installed and the R packages has been loaded. See below:
These are the list of required packages.
### Required R packages ----
bio_pkgs <- c("limma", "msigdbr", "DOSE", "devtools","fgsea", "enrichplot","GSVA","circlize",
"ComplexHeatmap","ggridges","pROC","cutpointr","ggalluvial","waterfalls",
"randomForest","devtools","tibble","msigdbr","data.table","tidyverse","dplyr",
"ggplot2","grid","gridExtra","ROCR","reshape2","stats","RColorBrewer","ggpubr",
"ggbeeswarm","tidyr","caret","escape", "doRNG")
github_pkgs <- c("kassambara/easyGgplot2")If not already installed, please do so as follows:
### If not already installed ----
suppressMessages({
if (!require(bio_pkgs, quietly = TRUE))
install.packages(bio_pkgs)
if (!require(github_pkgs, quietly = TRUE))
devtools::install_github(github_pkgs, dependencies = TRUE, force=TRUE)
})Lastly, load the R packages before you have your input data ready and run the function!
### Load R packages ----
suppressMessages(
lapply(bio_pkgs, library, character.only = TRUE)
)
suppressMessages(
library(easyGgplot2)
)The function outputs a list of results, including:
- Differential Gene Rankings (tabular format)
- Pairwise GSEA result (tabular format)
- Pairwise Barplot
- Pairwise Enrichment plots
- Single sample GSEA score result (tabular format)
- Single sample density plots
- Single sample histogram
- Single sample ROC plots
- Single sample cutoff labels (for ROC analysis)
- Single sample waterfall plots