To use the package, you first have to install it from GitHub using the
remotes package.
# Install remotes if not previously installed
if(!"remotes" %in% installed.packages()[,"Package"]) install.packages("remotes")
# Install rasterSp from Github if not previously installed
if(!"rasterSp" %in% installed.packages()[,"Package"]) remotes::install_github("RS-eco/rasterSp", build_vignettes = T)Load the rasterSp package
library(rasterSp)If you encounter a bug or if you have any problems, please file an issue on Github.
The rasterSp package includes various rasterized species data. For example, the bird distribution data can be loaded by:
data(ter_birds_dist)Note: The code for how this data was created data can be found in the data-raw folder.
filedir <- "/home/matt/Documents/"Note: You need to adapt filedir according to the path on your computer, where the IUCN Data is stored.
IUCN data was downloaded from: https://www.iucnredlist.org/resources/spatial-data-download, by running the following code:
if(!dir.exists(paste0(filedir, "/IUCN/TERRESTRIAL_MAMMALS"))){
download.file("http://spatial-data.s3.amazonaws.com/groups/TERRESTRIAL_MAMMALS.zip",
destfile = paste0(filedir, "/IUCN/TERRESTRIAL_MAMMALS.zip")) # <-- 594.3 MB
unzip(paste0(filedir, "/IUCN/TERRESTRIAL_MAMMALS.zip"), exdir = paste0(filedir, "/IUCN"))
unlink(paste0(filedir, "/IUCN/TERRESTRIAL_MAMMALS.zip"))
}You have one shapefile for a group of animals, consisting of individual polygons for each species.
Using the rasterizeIUCN function, the spatial distribution of each species is turned into a grid with a specific resolution. You can specify the resolution in degrees, here we use 0.5°. If save = TRUE, the grid for each species is saved to a GTiff file at the specified location (see filepath argument).
# Convert shape files into rasters and save to file
r_amphibians <- rasterizeIUCN(dsn=paste0(filedir, "/IUCN/AMPHIBIANS.shp"), resolution=0.5,
seasonal=c(1,2), origin=1, presence=c(1,2),
save=TRUE, path=paste0(filedir, "/SpeciesData/"))
# Is dropping 241 null geometries!!!
# Years are: 2008 2015 2009 2014 2013 2012 2010 2011 2016
r_odonata <- rasterizeIUCN(dsn=paste0(filedir, "/IUCN/FW_ODONATA.shp"), resolution=0.5,
seasonal=c(1,2), origin=1, presence=c(1,2),
save=TRUE, path=paste0(filedir, "/SpeciesData/"))
r_ter_mammals <- rasterizeIUCN(dsn=paste0(filedir, "/IUCN/TERRESTRIAL_MAMMALS.shp"), resolution=0.5,
seasonal=c(1,2), origin=1, presence=c(1,2),
save=TRUE, path=paste0(filedir, "/SpeciesData/"))
r_reptiles <- rasterizeIUCN(dsn=paste0(filedir, "/IUCN/Reptiles.shp"), resolution=0.5,
seasonal=c(1,2), origin=1, presence=c(1,2),
save=TRUE, path=paste0(filedir, "/SpeciesData/"))
# List of remaining IUCN files
species_groups <- c("CONESNAILS", "CORALS_PART_1", "CORALS_PART_2", "CORALS_PART_3", "LOBSTERS", "MANGROVES", "MARINEFISH_PART_1", "MARINEFISH_PART_2", "MARINEFISH_PART_3", "SEACUCUMBERS", "SEAGRASSES")
# Run rasterize on all species files
lapply(species_groups, FUN=function(x){
rasterizeIUCN(dsn=paste0(filedir, "/IUCN/", x, ".shp"),
resolution=0.5, save=TRUE, path=paste0(filedir, "/SpeciesData/"))})Bird data was obtained from BirdLife. Currently we use the 2014 version,
which comes in form of individual shapefiles for each species. Note:
The rasterizeIUCN function can also handle a list of shapefiles.
r_birds <- rasterizeIUCN(dsn=list.files(paste0(filedir, "/BirdLife/"), pattern=".shp", full.names=TRUE)[1:50],
resolution=0.5, save=TRUE, seasonal=c(1,2), origin=1, presence=c(1,2), df=TRUE,
path=paste0(filedir, "/SpeciesData/"))BirdLife as well as IUCN shapefiles provide information on a couple of
parameters (e.g. seasonal, origin and presence). These three parameters
are implemented in the rasterizeIUCN function, which then selects only
a specific subset of Polygons for each species. Infos on the different
parameters, can be found here:
http://datazone.birdlife.org/species/spcdistPOS.
The Birdlife 2017 data comes as gdb file, but can be converted in a shapfile by:
library(rgdal)
# The input file geodatabase
gdb_file = paste0(filedir, "BOTW/BOTW.gdb")
# List all feature classes in a file geodatabase
subset(ogrDrivers(), grepl("GDB", name))
fc_list = ogrListLayers(gdb_file)
print(fc_list)
# Read the feature class
botw <- readOGR(dsn=gdb_file, layer="All_Species")
# Save as shapefile
writeOGR(botw, dsn=paste0(filedir, "/BirdLife_2017/"), layer="All_Species", driver="ESRI Shapefile")In our case this was done in ArcMap, as it is much faster, although using sf might increase the performance in R considerably. The shapefile can then be loaded directly from file, by:
botw <- readOGR(dsn=paste0(filedir, "/BirdLife_2017/"), layer="All_Species")GARD reptile data was downloaded from: https://datadryad.org/resource/doi:10.5061/dryad.83s7k When using this data, please cite the original publication:
Roll U, Feldman A, Novosolov M, Allison A, Bauer AM, Bernard R, Böhm M, Castro-Herrera F, Chirio L, Collen B, Colli GR, Dabool L, Das I, Doan TM, Grismer LL, Hoogmoed M, Itescu Y, Kraus F, LeBreton M, Lewin A, Martins M, Maza E, Meirte D, Nagy ZT, de C. Nogueira C, Pauwels OSG, Pincheira-Donoso D, Powney GD, Sindaco R, Tallowin OJS, Torres-Carvajal O, Trape J, Vidan E, Uetz P, Wagner P, Wang Y, Orme CDL, Grenyer R, Meiri S (2017) The global distribution of tetrapods reveals a need for targeted reptile conservation. Nature Ecology & Evolution 1(11): 1677–1682. https://doi.org/10.1038/s41559-017-0332-2
Additionally, please cite the Dryad data package:
Meiri S, Roll U, Grenyer R, Feldman A, Novosolov M, Bauer AM (2017) Data from: The global distribution of tetrapods reveals a need for targeted reptile conservation. Dryad Digital Repository. https://doi.org/10.5061/dryad.83s7k
r_reptiles <- rasterizeIUCN(dsn=paste0(filedir, "/GARD1.1_dissolved_ranges/modeled_reptiles.shp"),
id="Binomial", split=NA, resolution=0.5, save=TRUE,
path=paste0(filedir, "/GARD_SpeciesData/"))The calcSR function uses a stepwise procedure to calculate the sum of species for each grid cell. This means only two files are loaded into memory at the same time to avoid memory shortage.
#Calculate amphibian richness
data(amphibians)
sr_amphibians <- calcSR(species_names=amphibians$binomial, path=paste0(filedir, "/SpeciesData/"))
raster::plot(sr_amphibians)However, this approach takes quite a while and means we have to recalculate the species richness everytime we change the species names. A much faster way is to save the presence of each species as a data frame and then only extract the species we are interested in. This is shown further below.
# Read species data
data("amphibians_dist")
data("odonata_dist")
data("ter_mammals_dist")
data("ter_birds_dist")
data("reptiles_dist")
data("gard_reptiles_dist")
# Create presence column
amphibians_dist$presence <- 1
odonata_dist$presence <- 1
ter_mammals_dist$presence <- 1
ter_birds_dist$presence <- 1
reptiles_dist$presence <- 1
gard_reptiles_dist$presence <- 1
# Create group column
amphibians_dist$group <- "Amphibians"
odonata_dist$group <- "Odonata"
ter_mammals_dist$group <- "Mammals"
ter_birds_dist$group <- "Birds"
reptiles_dist$group <- "Reptiles"
gard_reptiles_dist$group <- "GARD Reptiles"
# Calulate amphibian SR
library(dplyr)
sr_amphibians <- amphibians_dist %>% group_by(x, y, group) %>% summarise(sum = sum(presence))
# Calulate odonata SR
sr_odonata <- odonata_dist %>% group_by(x, y, group) %>% summarise(sum = sum(presence))
# Calulate terrestrial mammal SR
sr_ter_mammals <- ter_mammals_dist %>% group_by(x, y, group) %>% summarise(sum = sum(presence))
# Calulate terrestrial bird SR
sr_ter_birds <- ter_birds_dist %>% group_by(x, y, group) %>% summarise(sum = sum(presence))
# Calulate reptile SR
sr_reptiles <- reptiles_dist %>% group_by(x, y, group) %>% summarise(sum = sum(presence))
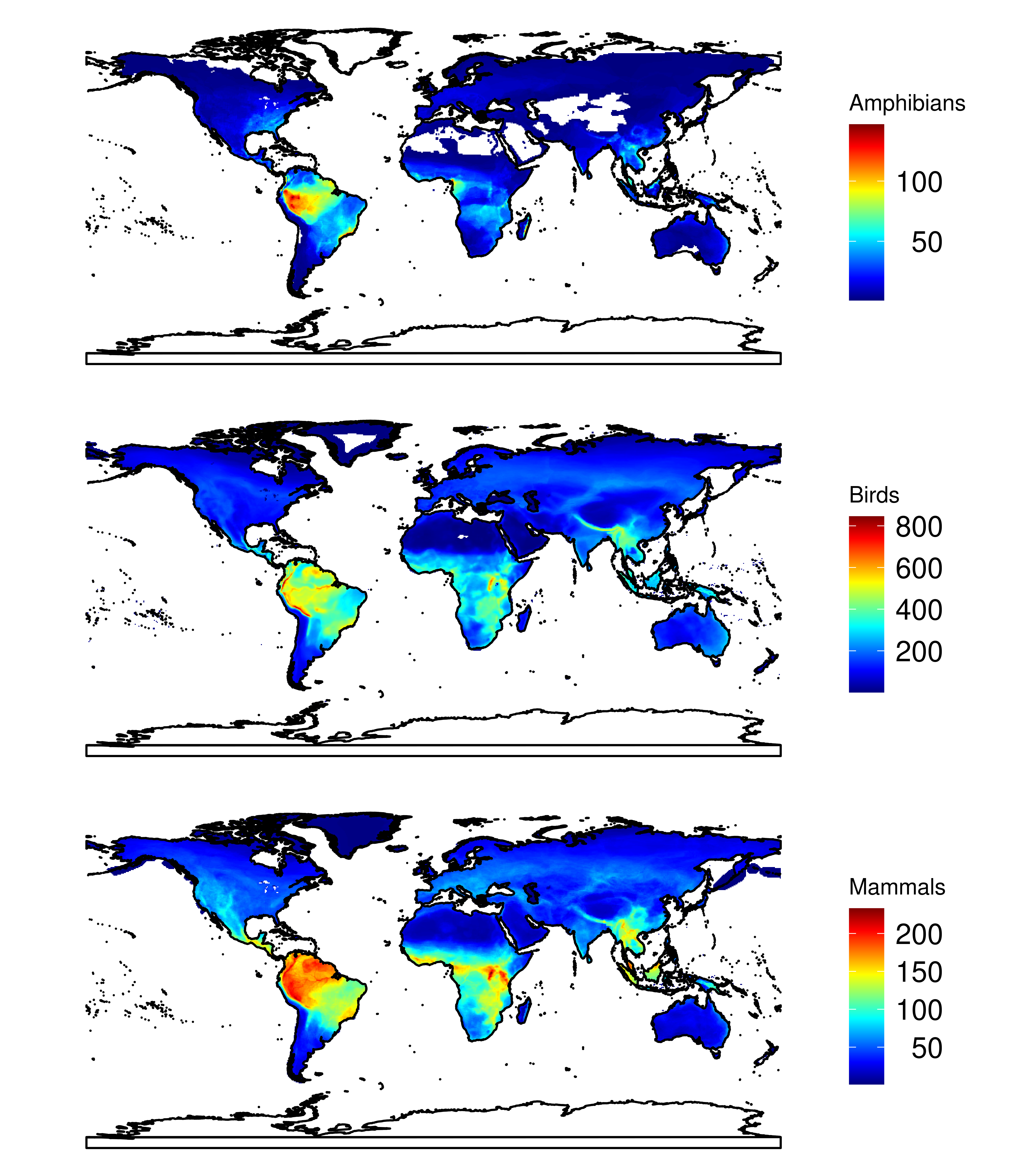
sr_gard_reptiles <- gard_reptiles_dist %>% group_by(x, y, group) %>% summarise(sum = sum(presence))library(ggmap2)
sr_alltaxa <- do.call(rbind, list(sr_amphibians, sr_ter_birds, sr_ter_mammals))
sr_alltaxa <- tidyr::spread(sr_alltaxa, group, sum)
ggmap2(sr_alltaxa, name=c("Amphibians", "Birds", "Mammals"), split=TRUE, ncol=1, country=T)library(ggmap2)
sr_rept <- do.call(rbind, list(sr_reptiles, sr_gard_reptiles))
sr_rept <- tidyr::spread(sr_rept, group, sum)
ggmap2(sr_rept, name=c("Reptiles", "GARD Reptiles"), split=TRUE, ncol=1, country=T)library(ggmap2)
sr_odonata <- tidyr::spread(sr_odonata, group, sum)
ggmap2(sr_odonata, name=c("Odonata"), ncol=1, country=T)Show distribution of number of presences per species
# Number of records per species
count_sp_amphibian <- amphibians_dist %>%
group_by(species, group) %>%
summarise(sum = sum(presence))
count_sp_odonata <- odonata_dist %>%
group_by(species, group) %>%
summarise(sum = sum(presence))
count_sp_ter_mammal <- ter_mammals_dist %>%
group_by(species, group) %>%
summarise(sum = sum(presence))
count_sp_ter_bird <- ter_birds_dist %>%
group_by(species, group) %>%
summarise(sum = sum(presence))
count_sp_reptiles <- reptiles_dist %>%
group_by(species, group) %>%
summarise(sum = sum(presence))
count_sp_gard_reptiles <- gard_reptiles_dist %>%
group_by(species, group) %>%
summarise(sum = sum(presence))
# Combine counts per species into one dataframe
species_presences_alltaxa <- rbind(count_sp_amphibian, count_sp_odonata, count_sp_ter_bird,
count_sp_ter_mammal, count_sp_reptiles, count_sp_gard_reptiles)
rm(count_sp_amphibian, count_sp_odonata, count_sp_ter_mammal, count_sp_ter_bird,
count_sp_reptiles, count_sp_gard_reptiles)Create Table with number of records per class and group
# Calculate number of records for each class (<10, 10-50, >50 records)
library(dplyr)
class1 <- species_presences_alltaxa %>% group_by(group) %>% filter(sum < 10) %>% summarise(n())
class2 <- species_presences_alltaxa %>% group_by(group) %>% filter(sum >= 10, sum <= 50) %>% summarise(n())
class3 <- species_presences_alltaxa %>% group_by(group) %>% filter(sum > 50) %>% summarise(n())
sum_records <- Reduce(function(x,y) merge(x,y, by="group"), list(class1, class2, class3))
colnames(sum_records) <- c("Group", "n < 10", "10 <= n <= 50", "n > 50")
# Create table
knitr::kable(sum_records, style="markdown")| Group | n < 10 | 10 <= n <= 50 | n > 50 |
|---|---|---|---|
| Amphibians | 3317 | 1471 | 1593 |
| Birds | 896 | 1549 | 7440 |
| GARD Reptiles | 2483 | 1953 | 3197 |
| Mammals | 968 | 1020 | 3288 |
| Odonata | 774 | 242 | 823 |
| Reptiles | 2047 | 1282 | 1970 |
#Calculate SR of for non-modelled species
library(dplyr)
sr_amphibians_smallrange <- amphibians_dist_smallrange %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
#sr_odonata_smallrange <- odonata_dist_smallrange %>%
# group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_ter_mammals_smallrange <- ter_mammals_dist_smallrange %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_ter_birds_smallrange <- ter_birds_dist_smallrange %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
#Create plot of global richness per group
sr_alltaxa_smallrange <- do.call(rbind, list(sr_amphibians_smallrange,
sr_ter_birds_smallrange,
sr_ter_mammals_smallrange))
sr_alltaxa_smallrange <- tidyr::spread(sr_alltaxa_smallrange, group, sum)
data(outline, package="ggmap2")
ggmap2::ggmap2(sr_alltaxa_smallrange, name=c("Amphibians", "Birds", "Mammals"),
split=TRUE, ncol=1, country=T)Plot SR of threatened species
#Calculate SR of for threatened species
library(dplyr)
sr_amphibians_threatened <- amphibians_threatened %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_ter_mammals_threatened <- ter_mammals_threatened %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_ter_birds_threatened <- ter_birds_threatened %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_alltaxa_threatened <- do.call(rbind, list(sr_amphibians_threatened,
sr_ter_birds_threatened,
sr_ter_mammals_threatened))
sr_alltaxa_threatened <- tidyr::spread(sr_alltaxa_threatened, group, sum)
library(ggmap2)
ggmap2(sr_alltaxa_threatened, name=c("Amphibians", "Birds", "Mammals"),
split=TRUE, ncol=1, country=T)Create dataframe of species data, which only occur in one country. And add country to species data.
#Rasterize country shapefile
data(countriesHigh, package="rworldxtra")
data(landseamask_generic, package="rISIMIP")
countries <- raster::rasterize(countriesHigh, landseamask_generic,
field="SOV_A3")
countries <- data.frame(raster::rasterToPoints(countries))
colnames(countries) <- c("x", "y", "country")
# Identify species that only occur in one country
amphibians_endemic <- amphibians_dist %>% left_join(countries) %>%
group_by(species) %>% summarise(n = n_distinct(country)) %>%
filter(n == 1)
ter_mammals_endemic <- ter_mammals_dist %>% left_join(countries) %>%
group_by(species) %>% summarise(n = n_distinct(country)) %>%
filter(n == 1)
ter_birds_endemic <- ter_birds_dist %>% left_join(countries) %>%
group_by(species) %>% summarise(n = n_distinct(country)) %>%
filter(n == 1)
# Subset species by endemism
amphibians_dist_endemic <- amphibians_dist %>%
filter(species %in% amphibians_endemic$species)
ter_mammals_dist_endemic <- ter_mammals_dist %>%
filter(species %in% ter_mammals_endemic$species)
ter_birds_dist_endemic <- ter_birds_dist %>%
filter(species %in% ter_birds_endemic$species)
# Calculate SR of endemic species
library(dplyr)
sr_amphibians_endemic <- amphibians_dist_endemic %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_ter_mammals_endemic <- ter_mammals_dist_endemic %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_ter_birds_endemic <- ter_birds_dist_endemic %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
# Plot SR of endemics
sr_alltaxa_endemic <- do.call(rbind, list(sr_amphibians_endemic,
sr_ter_birds_endemic,
sr_ter_mammals_endemic))
sr_alltaxa_endemic <- tidyr::spread(sr_alltaxa_endemic, group, sum)
library(ggmap2)
ggmap2(sr_alltaxa_endemic, name=c("Amphibians", "Birds", "Mammals"),
split=TRUE, ncol=1, country=T)# Load endemic species names
data(amphibians_endemic)
data(ter_mammals_endemic)
data(ter_birds_endemic)
# Calculate SR of endemic species
sr_amphibians_endemic <- amphibians_dist %>% filter(species %in% amphibians_endemic$species_name) %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_ter_mammals_endemic <- ter_mammals_dist %>% filter(species %in% ter_mammals_endemic$species_name) %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
sr_ter_birds_endemic <- ter_birds_dist %>% filter(species %in% ter_birds_endemic$species_name) %>%
group_by(x, y, group) %>% summarise(sum = sum(presence))
# Plot SR of endemics
sr_alltaxa_endemic <- do.call(rbind, list(sr_amphibians_endemic,
sr_ter_birds_endemic,
sr_ter_mammals_endemic))
sr_alltaxa_endemic <- tidyr::spread(sr_alltaxa_endemic, group, sum)
library(ggmap2)
ggmap2(sr_alltaxa_endemic, name=c("Amphibians", "Birds", "Mammals"), split=TRUE, ncol=1, country=T)To assess the invasiveness of species ranges, see the corresponding
vignette("iucn-invasions").