Sequence Validation Pipeline
- Download the entire repo (The Test_Scripts and MiSeqValidationResults folder are unnecessary)
- Install all the dependencies listed below
- Set paths in:
- pipeline.py => Set first 2
"System-Defined Variables"; Rest will be automatically set - PostProcessing/scripts/postanalysis/genedesign.py => Set
pathToPipeline="path/To/pipeline" - website/seqval.py => Set
pathToPipeline="path/To/pipeline"in functionrun_pipeline()
- pipeline.py => Set first 2
- Add JBEI SMB credentials to:
- MiSeqServerData.py => Everywhere there is the word
secret - website/seqval.py => Everywhere there is the word
secret
- MiSeqServerData.py => Everywhere there is the word
- Run it!
The website wil automatically be active at 127.0.0.1:82 To get the website interface running, run:
python seqval.py
Pipeline.py is a Python 3 script, so call it with Python 3
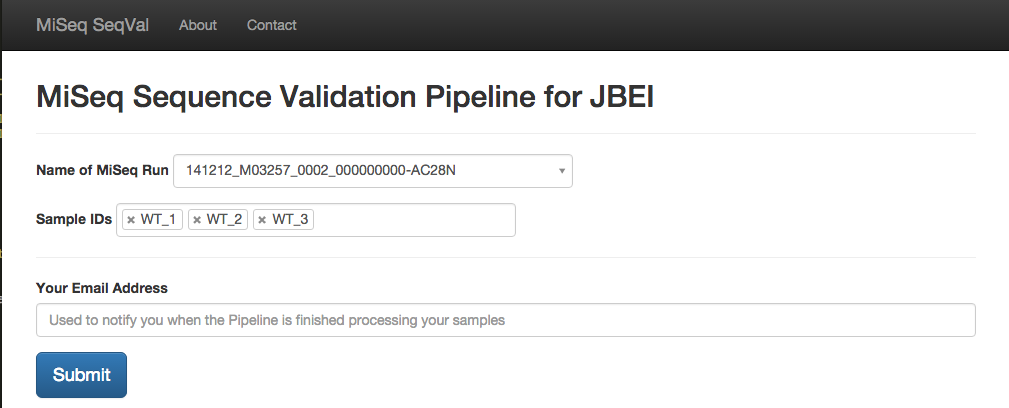
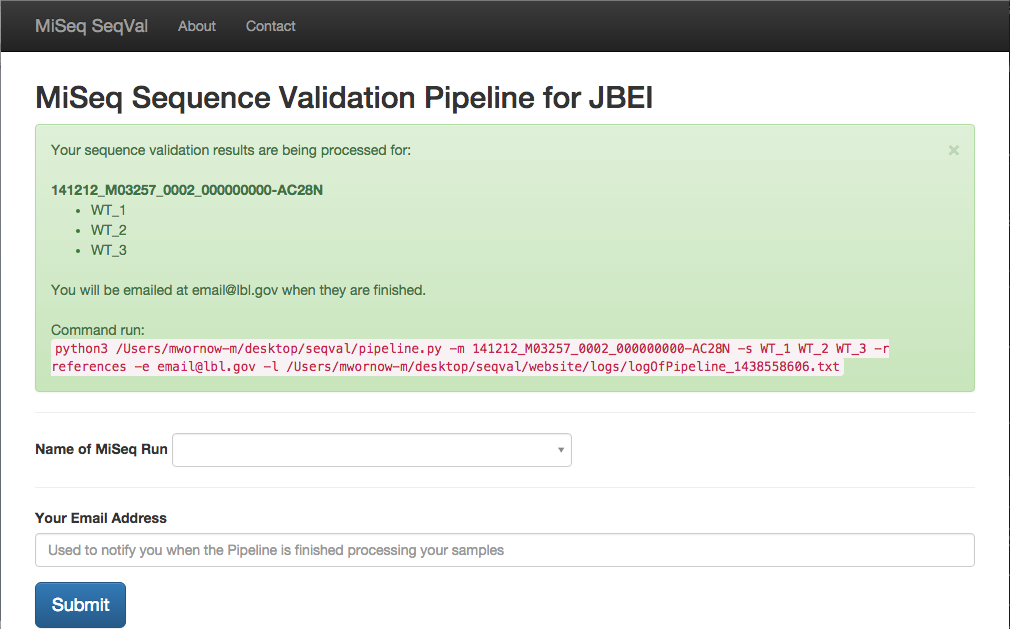
python3 pipeline.py -m <Main Library name> -s <Sub Library Names> -r <Reference sequence name> -e <Email address> -l <Path to logfile.txt> -n <Boolean, to use NERSC Version of tools>
- -m Main Library Name
- Name of Folder on MiSeq where this run is stored
- Example:
-m 141212_M03257_0002_000000000-AC28N
- -s Sub Library Names
- Space separated list of sample IDs that we want validated. These must be located in the Main Library folder
- Example:
-s WT_1 WT_2 WT_3 32_1
- -r Reference Sequences Names
- Space separated list of reference sequence names
- Example:
-r references1 references2
- -e Email Address
- Pipeline will send email to this address when Pipeline is finished
- Example:
-e email@lbl.gov
- -l Path to logfile.txt
- Path to where a Log File will be written after Pipeline is finished, detailing execution information
- Example:
-l /Users/synbio/sequencevalidation/logs
- -n Boolean flag for using NERSC version of tools
- If set, use the (older) NERSC Versions of the BWA, Samtools, and Picard tools
- Example:
-n
###Requirements:
-
Tools Folder (included with repo)
- GATK - 3.4-46
- BWA - 0.7.12
- Picard - 1.134
- Samtools - 1.12
- FastQC - 0.11.3
- DNAssemble - Perl Module
- BioDesign - Perl Module
-
Perl modules (Perl 5.18)
- Modern::Perl
sudo cpan install Modern::Perl
- Getup::Long
sudo cpan install Getup::Long
- Data::Alias
sudo cpan install Data::Alias
- Filename::Basename
sudo cpan install Filename::Basename
- Pod::Usage
sudo cpan install Pod::Usage
- List::MoreUtils
sudo cpan install List::MoreUtils
- YAML::Syck
sudo cpan install YAML::Syck
- Log::Log4perl
sudo cpan install Log::Log4perl
- File::NFSLock
sudo cpan install File::NFSLock
- Modern::Perl
-
Python
- lxml
pip install lxml
- scipy
pip install scipy
- numpy
pip install numpy
- openpyxl
pip install openpyxl
- biopython
pip install biopython
- pysmb
pip3 install pysmb
- flask (Only needed for Website)
pip install flask
- lxml
Note: Everything listed with just pip is installed for Python 2.7, since PostProcessing's scripts and Flask depend on Python 2.7.
- Other OS packages:
- zlib
sudo apt-get install zlib
- curses
sudo apt-get install libncurses5-dev
- pigz
sudo apt-get install pigz
- Java Runtime Environment
sudo apt-get install default-jre
- zlib
- Take MiSeq files from the SMB server
- Create .bam files
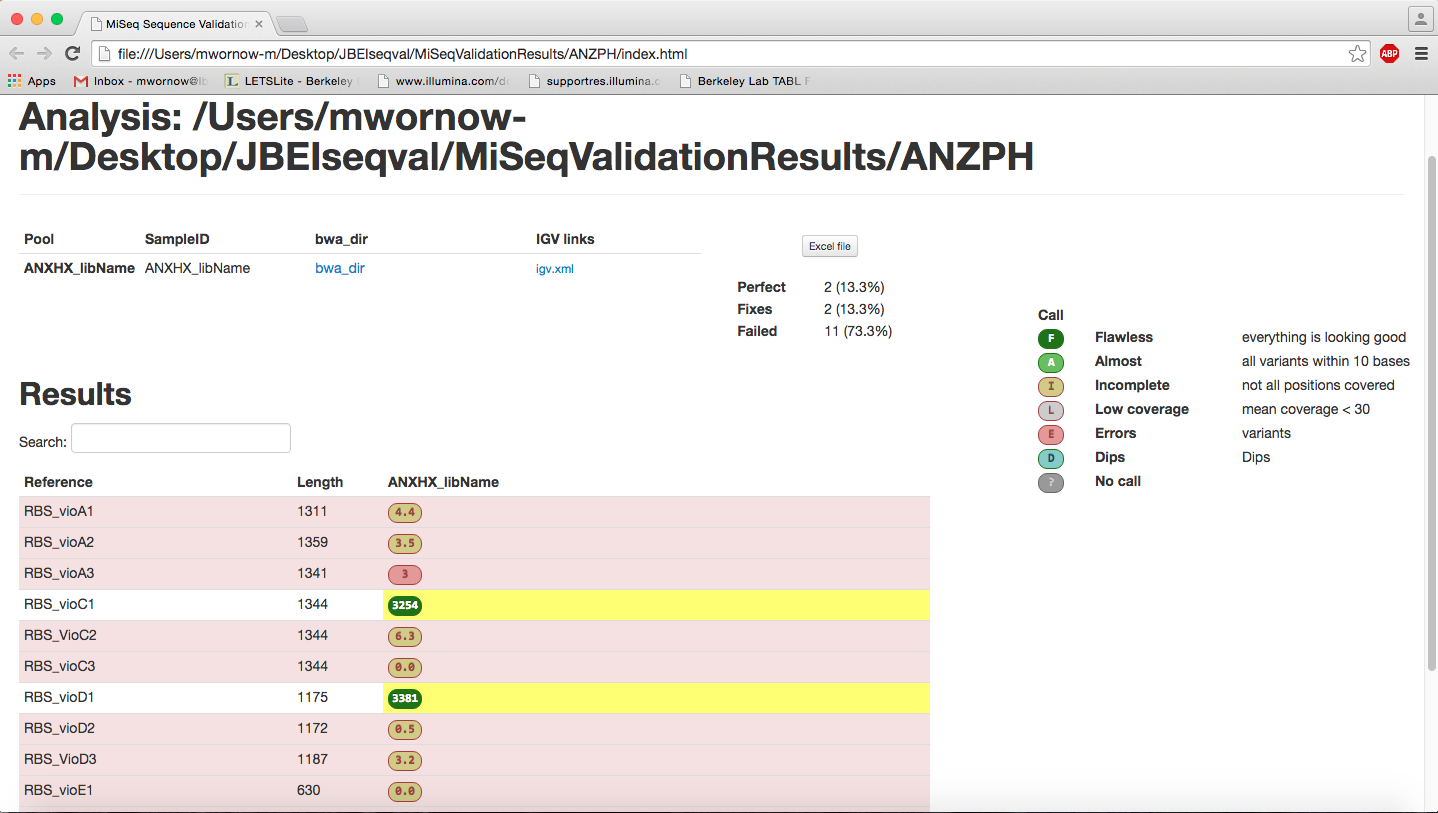
- Use GATK to generate coverage depth, .vcf, .bed, and call_summary.txt
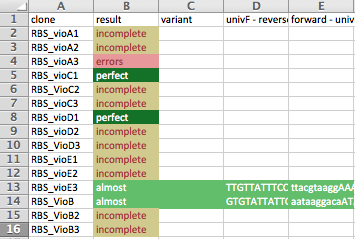
- Make calls and generate summary: IGV.xml, Excel, and HTML
Special thanks to Ernst, Joel, and Oge!
See Presentation/Presentation.pptx for more!