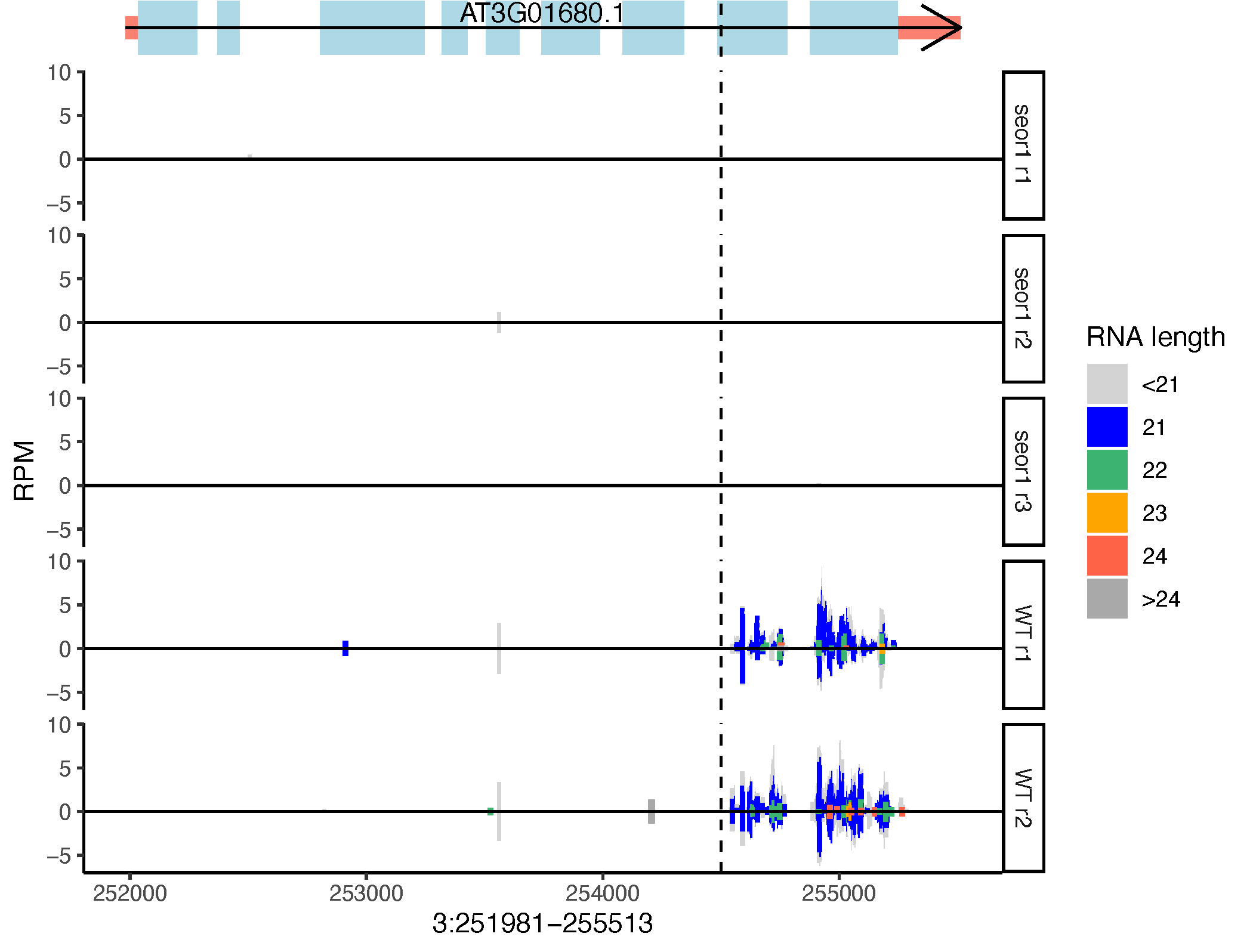
Visualization of plant smallRNA-seq alignments, with an emphasis on distinguishing biologically relevant differences in small RNA size
Michael J. Axtell, The Pennsylvania State University, USA mja18@psu.edu
Genome browsers are ill-suited to render alignments of small RNA-seq data. To interpret this type of data, users to need to be able to visualize three parameters:
- Depth of coverage
- Strandedness of coverage
- Small RNA sizes
Most (all?) genome browsers cannot simultaneously visualize all three of these track parameters. Furthermore, users often want to compare multiple tracks visually. This requires a set normalization (such as reads per million) be applied to each alignment track AND the y-axis scaling set in a uniform fashion across all tracks. Genome browsers generally can't do this.
So, this script is a way to visualize small RNA-seq alignments that does meet all of these parameters.
- Linux or Mac OS
- R , with packages "tidyverse", "cowplot", "IRanges" and "docopt" installed.
- samtools , installed in the user's PATH. Requires samtools version >= 1.10 !
- Download script and
chmod +x sRNA_Viewerto make the script executable. If desired move to a location in yourPATH.
NOTE : sRNA_Viewer uses a shebang on the first line to specify #!/usr/bin/env Rscript --vanilla. If your system has Rscript somewhere else, you will need to explicitly call Rscript --vanilla sRNA_Viewer ....
sRNA_Viewer [-g TABIXGFF -l VLINE -r RGLIST] -c COORDINATES -b BAMLIST -p OUTPUTFILE
sRNA_Viewer -h | --help
sRNA_Viewer -v | --version
-c COORDINATES: Location to analyze in format Chr:Start-Stop (one-based, inclusive)-b BAMLIST: A .csv file listing the bamfiles, along with the user's preferred display names. The display names are what each track will be labeled with on the figure (unless read-groups are used, see below). Each bam file must be readable, positionally sorted, and already indexed (usingsamtools index).-p OUTPUTFILE: Output .pdf file to be created.
-g TABIXGFF: tabix-ed gff3 file containing transcript/mRNA, exon, and CDS. See section below for preparation instructions. Providing this file will allow rendering of transcript models.-l VLINE: x-coordinate to draw a vertical line at. A single integer, within the coordinates provided by option-c.-r RGLIST: This is a csv file with read groups in column 1, display names in column 2. Applies to first bamfile from BAMLIST. Providing this information will cause the specified read groups (specified by @RG tags) to be plotted in separate tracks. This will applied to the first bam file in the-b BAMLISTfile only.
sRNA_Viewer can use a bgzip-compressed, tabix-indexed GFF3 file of transcripts. Only the annotation types of transcript, mRNA, exon, and CDS are recognized and used.
Here is an example of how to process a gff3 file for use by sRNA_Viewer
grep -e '\smRNA\s' -e '\stranscript\s' -e '\sexon\s' -e '\sCDS\s' mygff.gff | sort -k1,1 -k4,4n | bgzip > mygff_sorted.gff.gz
tabix -p gff mygff_sorted.gff.gz
This will create two new files: mygff_sorted.gff.gz and its index mygff_sorted.gff.gz.tbi. In option -g specify the file mygff_sorted.gff.gz. (The corresponding index must be in the same location).
Using only -b to specify multiple individual bam files is faster than using -b and -r to specify multiple read-groups from a single merged bam file.