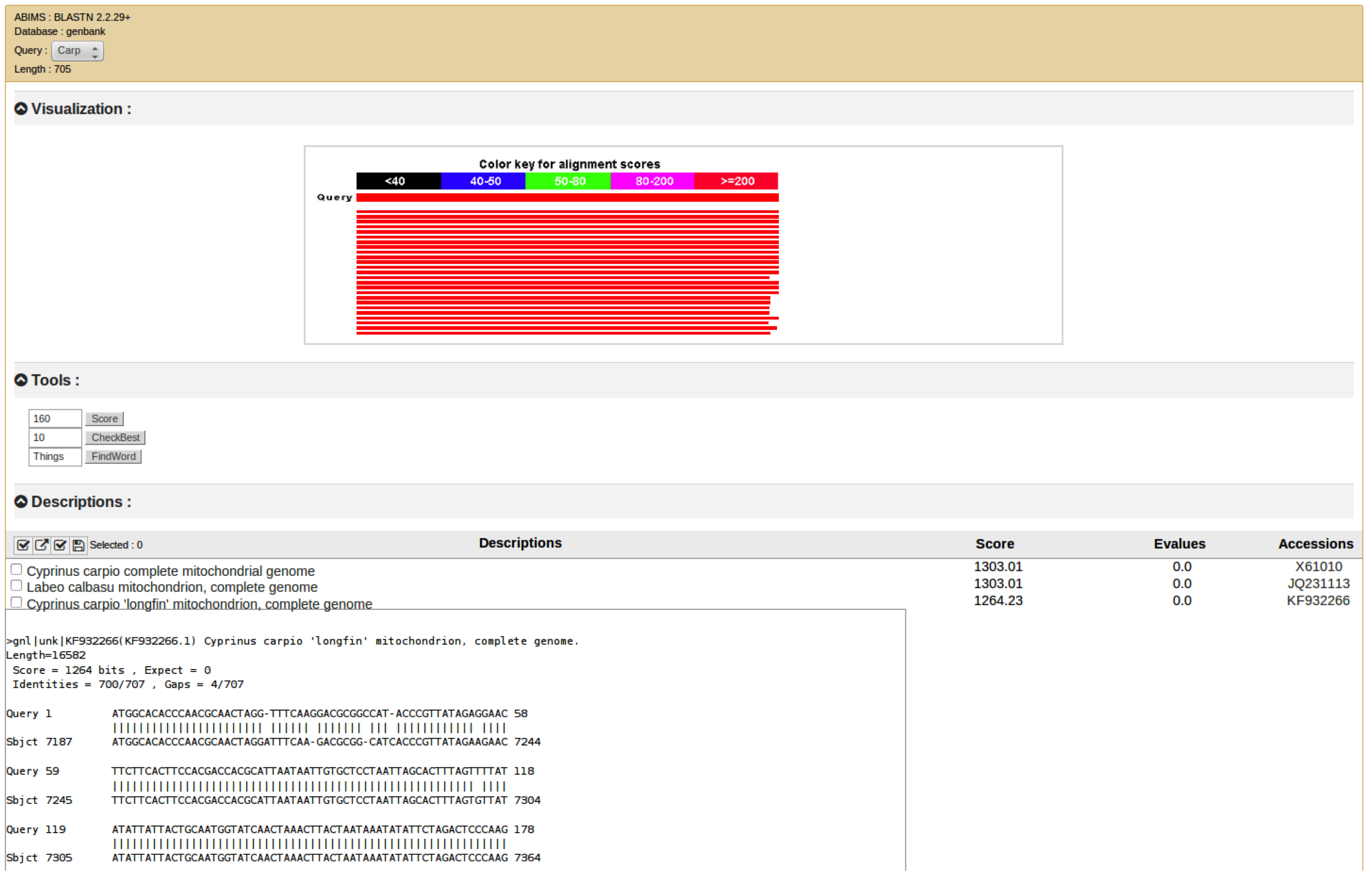
Blastvisu is a project support by ABiMS It aim to provide the equivalent of NCBI-W3BLAST and NCBI-BLAST on Galaxy.
git clone https://github.com/lecorguille/galaxy-blastvisu.git
cp -r galaxy-blastvisu/plugin $GALAXY_ROOT/config/plugins/visualization/blastvisu
cp -r galaxy-blastvisu/static/visuBlast $GALAXY_ROOT/static
Restart Galaxy
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families.
Source: http://blast.ncbi.nlm.nih.gov/Blast.cgi
Galaxy is an open, web-based platform for data intensive biomedical research. Whether on the free public server or your own instance, you can perform, reproduce, and share complete analyses.
Homepage: https://galaxyproject.org/
The visualisation is design to display the XML format which can be produce by the @peterjc tools Galaxy wrappers for NCBI BLAST+ and related BLAST tools
- Emma Prudent - ABiMS / IFB - UPMC/CNRS - Station Biologique de Roscoff - France
- Caroline Vernette - ABiMS / IFB - UPMC/CNRS - Station Biologique de Roscoff - France
- Gildas Le Corguillé @lecorguille - ABiMS / IFB - UPMC/CNRS - Station Biologique de Roscoff - France