This fork has edits made in-place for allowing direct training. Also includes notebook (recommended Colab) for step-by-step approach. The model includes 'Dropout' layer. Feel free to test out your own variants in architecture.
To download the dataset zip, head to Penn Imaging Home and apply for permissions. Paste the Training Data link at the appropriate cell in the notebook.
Other notes:
- To amend your model/training params/checkpoint creation, download the necessary
.pyscript from colab, update and use!mv src destto apply changes. - The script by default, saves model after every epoch.
extract_patches.pyserves the purposes for both training and validation data generation.- To generate validation data, you could extract a portion of the training numpy array that is generated by
extract_patches.py. Not much edits intrain.pyandextract_patches.pyare needed.
This repository provides source code for a deep convolutional neural network architecture designed for brain tumor segmentation with BraTS2017 dataset. The architecture is fully convolutional network (FCN) built upon the well-known U-net model and it makes use of residual units instead of plain units to speedup training and convergence. The Brain tumor segmentation problem exhibits severe class imbalance where the healthy voxels comprise 98% of total voxels,0.18% belongs to necrosis ,1.1% to edema and non-enhanced and 0.38% to enhanced tumor. The issue is addressed by: 1) adopting a patch-based training approach; 2) using a custom loss function that accounts for the imbalance. During training, 2D patches of size 128x128 from the axial plane are randomly sampled. And by doing so it allows to dismiss patches from pixels with zero intensity and therefore it helps a bit to alleviate the problem.
The implementation is based on keras and tested on both Theano and Tensorflow backends.
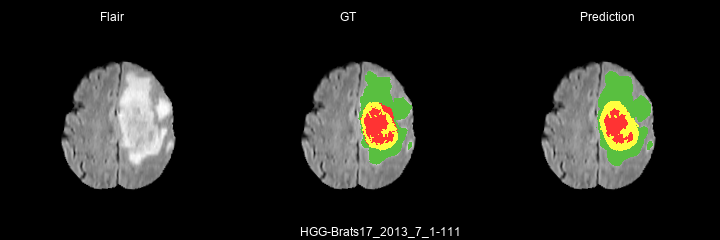
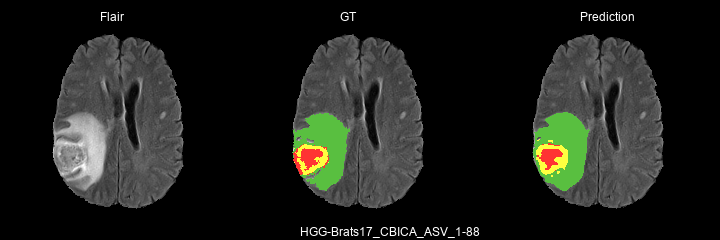
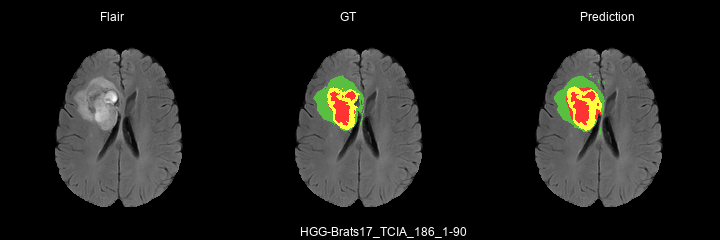
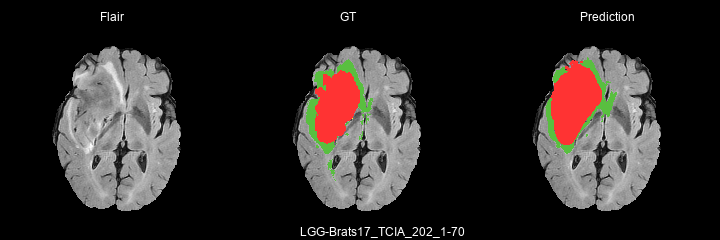
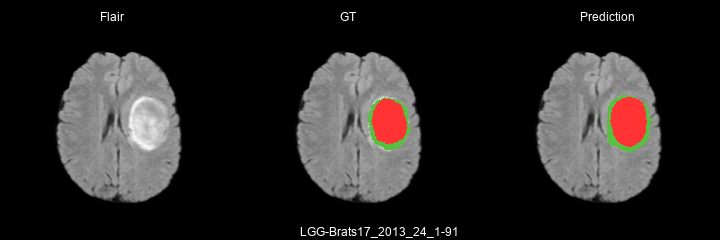
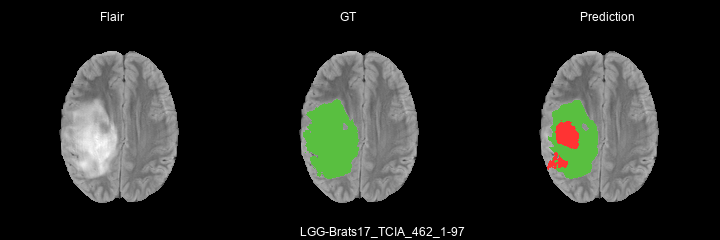
Here are some results predicted by a model trained for 2 epochs :
- HGG cases :
- LGG cases :
To run the code, you first need to install the following prerequisites:
- Python 3.5 or above
- numpy
- keras
- scipy
- SimpleITK
- Execute first
extract_patches.pyto prepare the training and validation datasets. - then
train.pyto train the model. predict.pyto make final predictions.
python extract_patches.py
python train.py
python predict.py
This code is an implementation of this paper. If you find this code useful in your research, please consider citing:
@inproceedings{kermi2018deep,
title={Deep Convolutional Neural Networks Using U-Net for Automatic Brain Tumor Segmentation in Multimodal MRI Volumes},
author={Kermi, Adel and Mahmoudi, Issam and Khadir, Mohamed Tarek},
booktitle={International MICCAI Brainlesion Workshop},
pages={37--48},
year={2018},
organization={Springer}
}