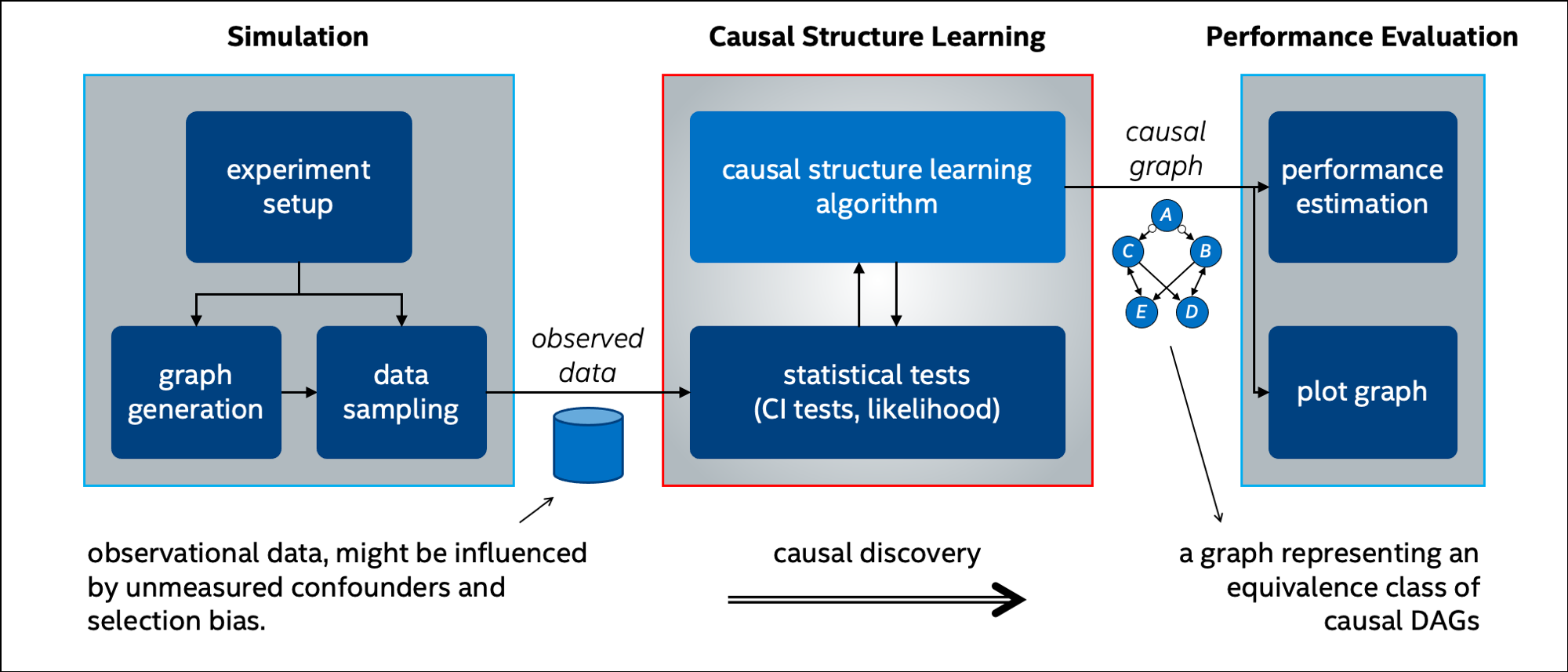
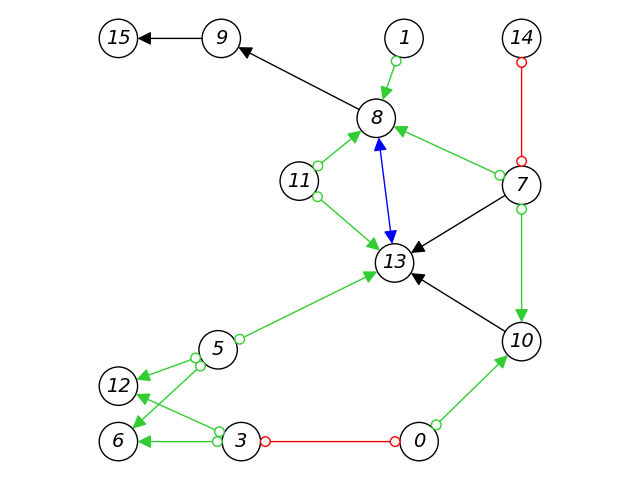
This repository contains research code of novel causal discovery algorithms developed at Intel Labs, as well as other common algorithms, and classes for developing and examining new algorithms for causal structure learning.
Included algorithms learn causal structures given observed data. They assume causal Markov condition and faithfulness. There are two families of algorithms:
-
Causal discovery under causal sufficiency and bayesian network structure learning
- PC algorithm (Spirtes et al., 2000)
- RAI algorithm, Recursive Autonomy Identification (Yehezkel and Lerner, 2009). This algorithm is used for learning the structure in the B2N algorithm (Rohekar et al., NeurIPS 2018b)
- B-RAI algorithm, Bootstrap/Bayesian-RAI for uncertainty estimation (Rohekar et al., NeurIPS 2018a). This algorithm is used for learning the structure of BRAINet (Rohekar et al., NeurIPS 2019)
-
Causal discovery in the presence of latent confounders and selection bias
- FCI algorithm, Fast Causal Inference (Spirtes et at., 2000)
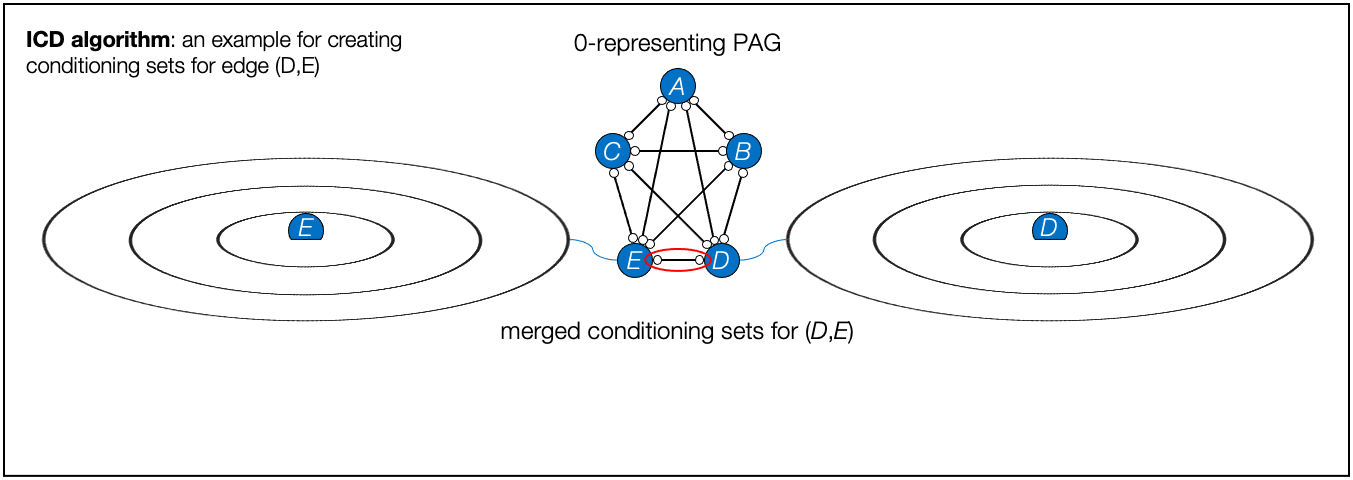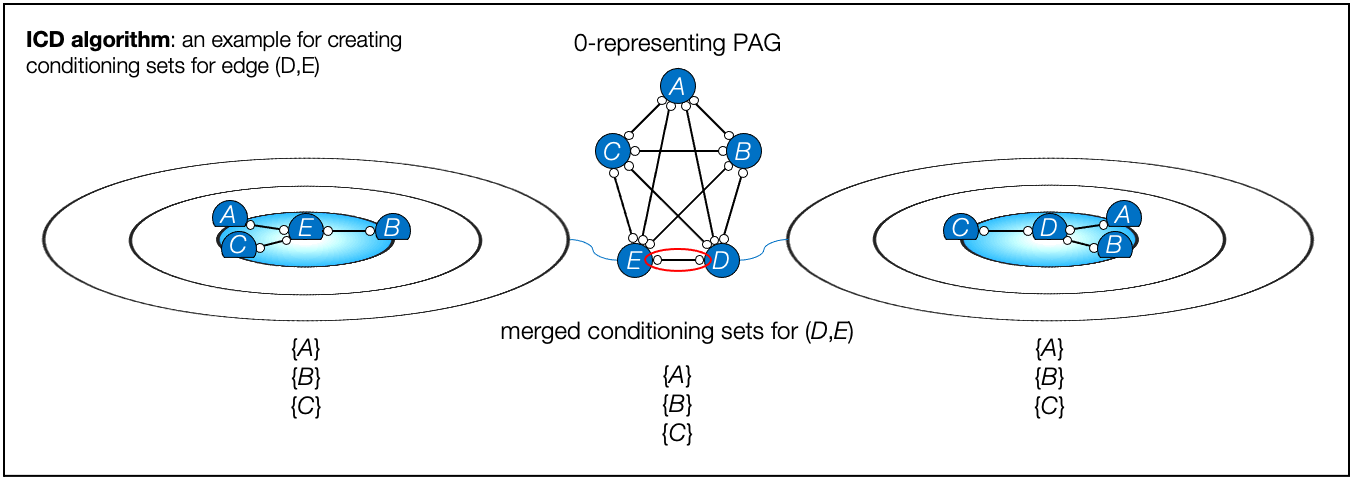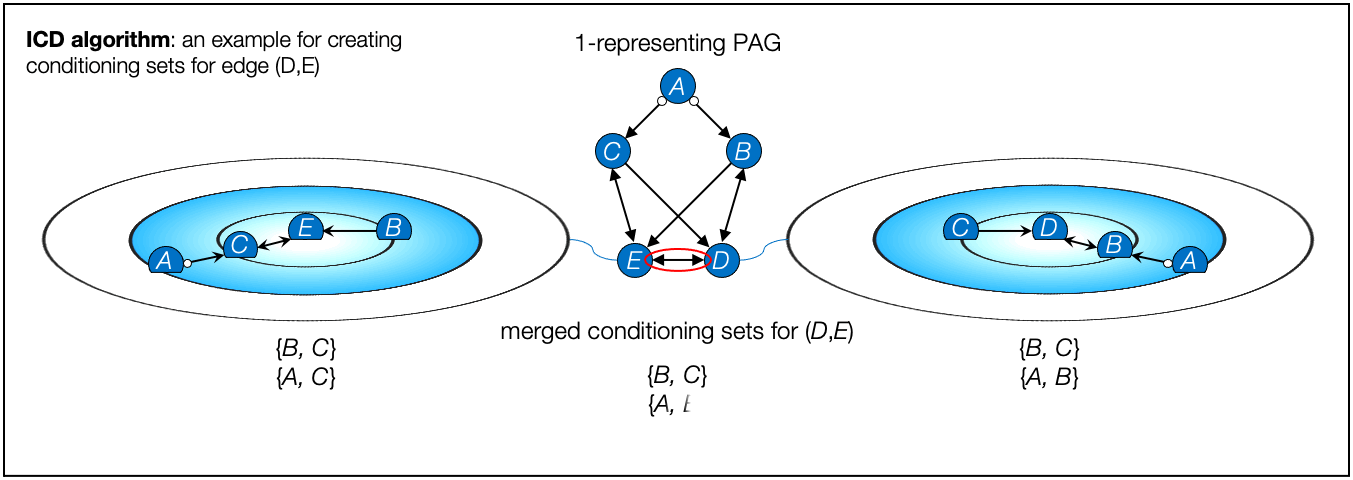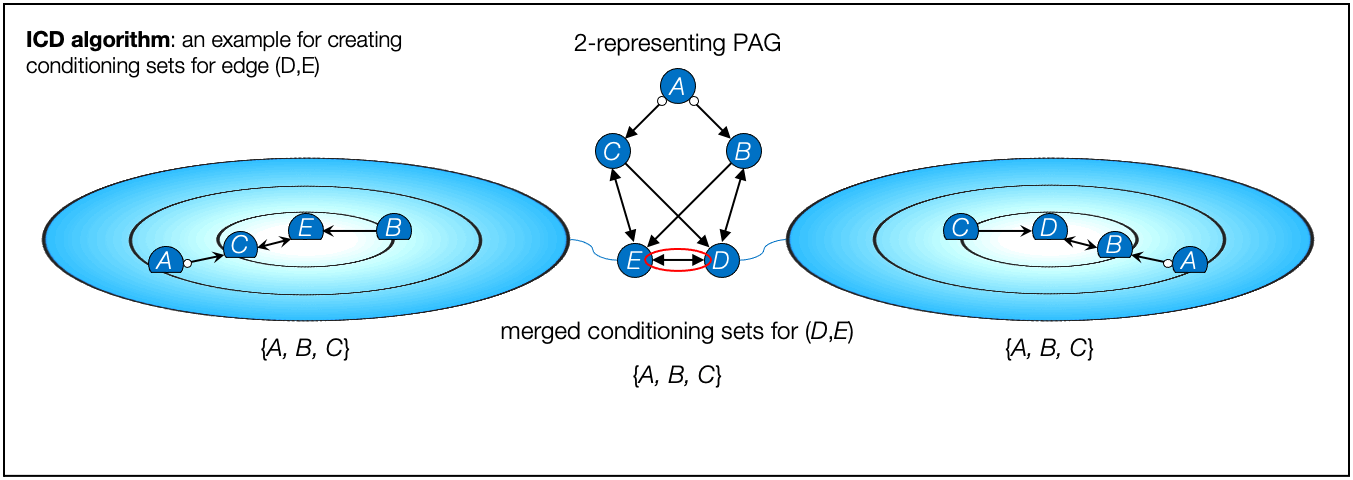
- ICD algorithm, Iterative Causal Discovery (Rohekar et al., NeurIPS 2021)
This repository includes several classes and methods for implementing new algorithms and testing them. These can be grouped into three categories:
- Simulation:
- Causal structure learning:
- Classes for handling graphical models (e.g., methods for graph traversal and calculating graph properties). Supported graph types:
- Directed acyclic graph (DAG): commonly used for representing causal DAGs
- Partially directed graph (PDAG/CPDAG): a Markov equivalence class of DAGs under causal sufficiency
- Undirected graph (UG) usually used for representing adjacency in the graph (skeleton)
- Ancestral graph (PAG/MAG): a MAG is an equivalence class of DAGs, and a PAG is an equivalence class of MAGs (Richardson and Spirtes, 2002).
- Statistical tests (CI tests) operating on data and a perfect CI oracle (see causal discovery with a perfect oracle)
- Classes for handling graphical models (e.g., methods for graph traversal and calculating graph properties). Supported graph types:
- Performance evaluations:
- Graph structural accuracy
- Skeleton accuracy: FNR, FPR, structural Hamming distance
- Orientation accuracy
- Overall graph accuracy: BDeu score
- Computational cost: Counters for CI tests (internal caching ensures counting once each a unique test)
- Plots for DAGs and ancestral graphs.
- Graph structural accuracy
A new algorithm can be developed by inheriting classes of existing algorithms (e.g., B-RAI inherits RAI) or by creating a new class.
The only method required to be implemented is learn_structure(). For conditional independence testing,
we implemented conditional mutual information, partial correlation statistical test, and d-separation (perfect oracle).
Additionally, a Bayesian score (BDeu) can be used for evaluating the posterior probability of DAGs given data.
This code has been tested on Ubuntu 18.04 LTS and macOS Catalina, with Python 3.5. We recommend installing and running it in a virtualenv.
sudo -E pip3 install virtualenv
virtualenv -p python3 causal_env
. causal_env/bin/activate
git clone https://github.com/IntelLabs/causality-lab.git
cd causality-lab
pip install -r requirements.txt
All causal structure learning algorithms are classes with a learn_structure() method that learns the causal graph.
The learned causal graph is a public class member, simply called graph, which is an instance of a graph class.
The structure learning algorithms does not have direct access to the data, instead they call a statistical test which accesses the data.
Let's look at the following example: causal structure learning with ICD using a given dataset.
par_corr_test = CondIndepParCorr(dataset, threshold=0.01) # CI test with the given significance level
icd = LearnStructICD(nodes_set, par_corr_test) # instantiate an ICD learner
icd.learn_structure() # learn the causal graph
For complete examples, see causal discovery with latent confounders and causal discovery under causal sufficiency notebooks. The learned structures can then be plotted - see a complete example for creating a PAG, calculating its properties, and plotting it in the partial ancestral graphs notebook.
- Richardson, Thomas, Peter Spirtes. "Ancestral graph Markov models". The Annals of Statistics, 30 (4): 962–1030, 2002.
- Rohekar, Raanan Y., Yaniv Gurwicz, Shami Nisimov, Guy Koren, and Gal Novik. "Bayesian Structure Learning by Recursive Bootstrap." Advances in Neural Information Processing Systems 31: 10525-10535, 2018.
- Rohekar, Raanan Y., Shami Nisimov, Yaniv Gurwicz, Guy Koren, and Gal Novik. "Constructing Deep Neural Networks by Bayesian Network Structure Learning". Advances in Neural Information Processing Systems 31: 3047-3058, 2018b.
- Rohekar, Raanan Y., Yaniv Gurwicz, Shami Nisimov, and Gal Novik. "Modeling Uncertainty by Learning a Hierarchy of Deep Neural Connections". Advances in Neural Information Processing Systems 32: 4244-4254, 2019.
- Rohekar, Raanan Y., Shami Nisimov, Yaniv Gurwicz, and Gal Novik. "Iterative Causal Discovery in the Possible Presence of Latent Confounders and Selection Bias." Advances in Neural Information Processing Systems 34, 2021.
- Spirtes, Peter, Clark N. Glymour, Richard Scheines, and David Heckerman. "Causation, prediction, and search". MIT press, 2000.
- Yehezkel, Raanan, and Boaz Lerner. "Bayesian Network Structure Learning by Recursive Autonomy Identification". Journal of Machine Learning Research 10, no. 7, 2009