Source code for our paper: "GraphCare: Enhancing Healthcare Predictions with Open-World Personalized Knowledge Graphs".
Requirements:
pip install torch==1.12.0
pip install torch-geometric==2.3.0
pip install pyhealth==1.1.2
pip install scikit-learn==1.2.1
pip install openai==0.27.4We follow the flow of methodology section (Section 3) to explain our implementation.
The jupyter notebook to prompt KG for EHR medical code:
/graphcare_/graph_generation/graph_gen.ipynbWe place sample KGs generated by GPT-4 as
/graphs/{condition/CCSCM,procedure/CCSPROC,drug/ATC3}/{code_id}.txtThe script for subgraph sampling from UMLS:
/KG_mapping/umls_sampling.pyWe place 2-hop sample KGs randomly subsampled from UMLS as
/graphs/umls_2hop.csvThe jupyter notebooks for word embedding retrieval:
/graphcare_/graph_generation/{cond,proc,drug}_emb_ret.ipynbDue to the large size of word embedding, we do not include them in the repo. You can use our script to retrieve it and store it in either
/graphs/cond_proc/{entity_embedding.pkl, relation_embedding.pkl}
or
/graphs/cond_proc_drug/{entity_embedding.pkl, relation_embedding.pkl}depending on the features used for the prediction tasks.
The function for node & edge clustering:
clustering() in data_prepare.pyWe place some clustering results (only "_inv" as cluster embedding has large size) in
/clustering/process_sample_dataset() and process_graph() in data_prepare.py
&
get_subgraph() in graphcare.pyThe implementation of our proposed BAT model is in
/graphcare_/model.pyThe creation of task-specific datasets (using PyHealth) is in
data_prepare.pyThe training and prediction details are in
graphcare.pyThe scripts running baseline models are placed in
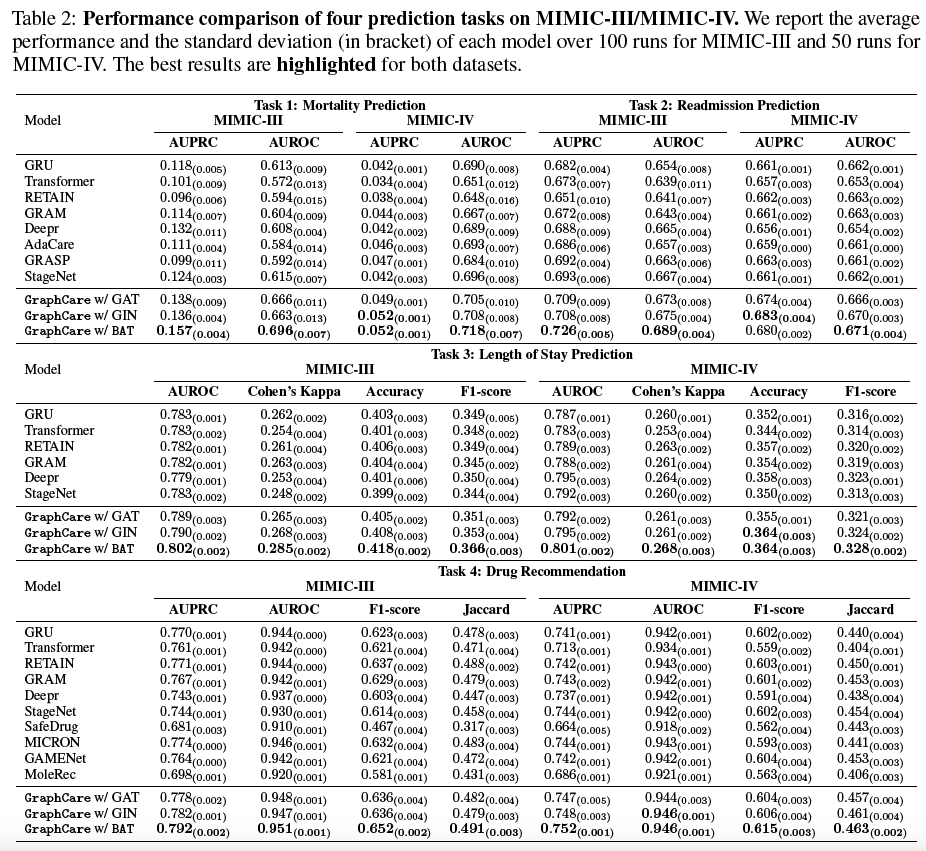
/baselinesWe show precise (three decimal digits) results (equivalent to Table 2 in the paper) as follows: